
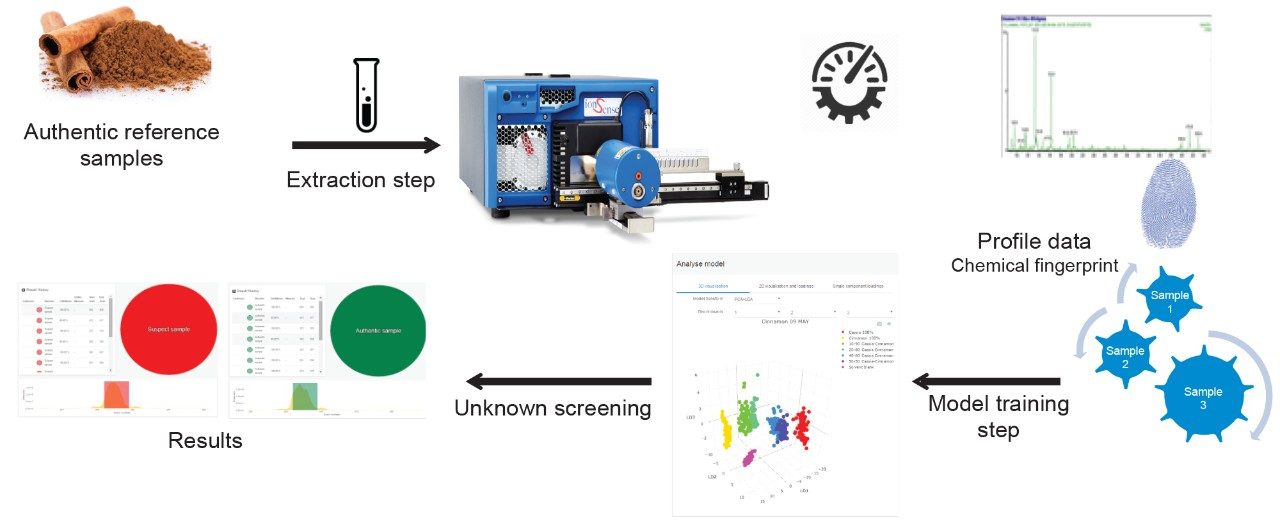
In this application note, we demonstrate the use of DART coupled to Waters ACQUITY QDa Mass Detector with chemometric modeling performed in real time with LiveID Software to rapidly determine the species level identity of cinnamon for label claim verification and authenticity purposes for the food industry.
Cinnamon is a popular, aromatic culinary spice; it is produced from the inner bark of the Cinnamomum tree which belongs to the genus Cinnamomum of the Laurel family (Lauraceae).
Cinnamon has many species that differ in smell, taste, and color depending on the geographical region, but the species of most commercial importance are Cinnamomum verum and Cinnamomum cassia. Cinnamomum verum is known as Ceylon cinnamon or “true cinnamon” while C. cassia is the Chinese species. Both species have similar characteristics that exhibit a fragrant, sweet, and warm taste; however, the flavor of the Ceylon variety is more refined and subtle.1 The unique properties of this spice come from its essential oils and compounds, in particular cinnamaldehyde. Cinnamaldehyde is the compound which gives cinnamon its flavor and aroma, and it is also proposed to be responsible for many of the health benefits associated with cinnamon ingestion. Strips of the inner bark are dried until they curl into rolls known as cinnamon sticks or quills. These can then be further ground into powder or made into an extract.
C. cassia is economically cheaper and more abundant than the C. verum species with its value depending on the percentage of cinnamaldehyde. C. cassia is also known to contain coumarin, cinnamyl acetate, cinnamic acid, phenylpropyl acetate, orthocumaric aldehyde, and tannic acid. The most common form of cinnamon adulteration is via the substitution or dilution of ground C. verum with C. cassia. European health agencies have warned against consuming high amounts of C. cassia due to the elevated amount of coumarin, a known blood-thinning agent which could damage the liver if taken in large amounts. Other bioactive compounds found in the bark, powder, and essential oils of C. cassia are cinnamaldehyde and styrene. In high doses these substances can also be toxic for humans.2
The IonSense Direct Analysis in Real Time (DART) is a form of ambient mass spectrometry which is defined as mass spectrometric analysis with minimal effort for sample preparation, using direct sampling and ionization at ambient conditions.3 DART is an atmospheric pressure ionization related technique based on the thermal desorption of condensed phase analytes by a plasma discharge in a heated gas stream, typically helium or nitrogen. Metastable atoms generated from gas interact with ambient molecules, such as water to create gas-phase ionic reagents which in turn react and ionize analytes on a surface, or present as a vapor in the atmosphere. DART is capable of analyzing low to high polarity compounds (up to 1 kDa) in both negative ion and positive ion modes.
In this application note, we demonstrate the use of DART coupled to Waters ACQUITY QDa Mass Detector with chemometric modeling performed in real time with LiveID Software to rapidly determine the species level identity of cinnamon for label claim verification and authenticity purposes for the food industry.
The chemometric model was trained using authentic ground samples of Ceylon cinnamon (C. verum) and C. cassia obtained from a national herb and spices supplier. Authentic samples of Ceylon cinnamon were mixed in variant percentage with C. cassia to investigate the capability of the method to detect mixtures of the cinnamon species in the case of adulteration by substitution or dilution.
To detect compounds of interest 1 g homogenized sample of ground cinnamon/cassia was weighed into a 50-mL tube and mixed with 15 mL of EtOH:water 50:50 (v/v). The sample was mixed using vortex mixer for 30 s followed by sonication for 15 min. The samples were then centrifuged at 6000 rpm for 5 min, and 3 mL of supernatant was loaded onto an OASIS HLB Cartridge, 3 cc, 60 mg (p/n: WAT094226). Elution was performed using 1 mL of methanol. Two replicate extracts were prepared for each sample and stored at -20 °C. Prior to analysis, the extracts were spotted (3 µL) on the 12 positions of a QuickStrip card and allowed to dry under ambient conditions. Each QuickStrip card was then analyzed using DART QDa, thus generating 12 replicate Regions of Interest (ROIs) per card, per extract.
|
MS system: |
ACQUITY QDa |
|
MS source: |
DART |
|
Ionization mode: |
Positive |
|
Acquisition mode: |
Full scan MS |
|
Gas temp. (He): |
150 °C |
|
Sampling speed: |
1.00 mm/sec |
|
Sampling frequency: |
2 Hz |
|
Cone voltage: |
10 V |
|
Mass range: |
100–600 m/z (continuum) |
LiveID multivariate statistical software package (v1.2) was used as a chemometric model building and real time recognition tool.
The workflow illustrated in Figure 1 was followed for chemometric model training.
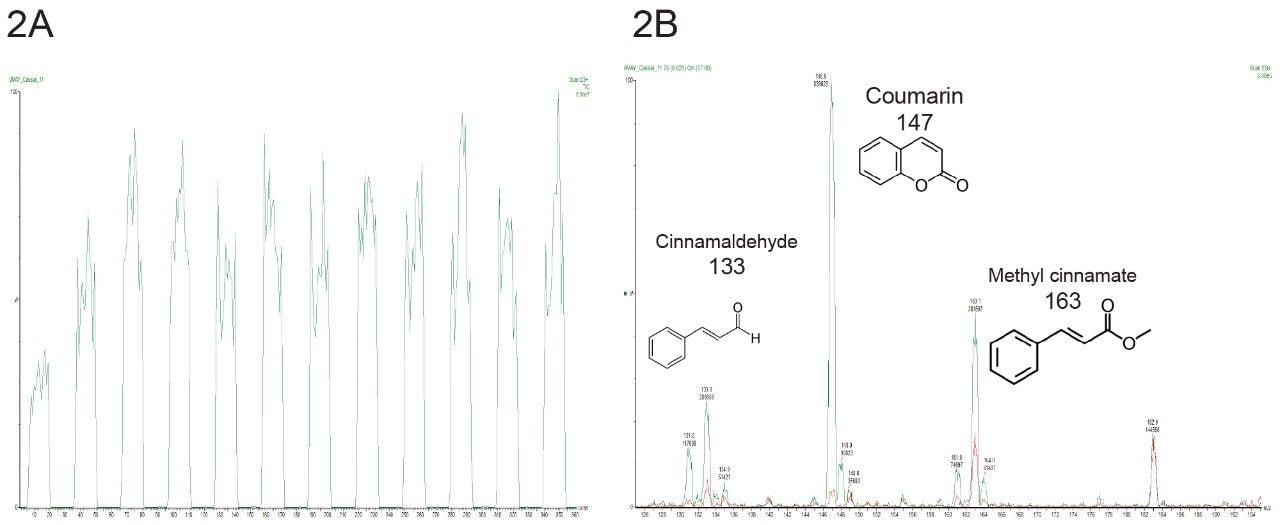
Combined spectrometric data (Figure 2) obtained from two different production lots of authentic C. verum (n=59) and C. cassia (n=60), and mixtures of the two species at portions representing 50:50% (n=63) and 10:90% (n=74) were used to train the chemometric model.
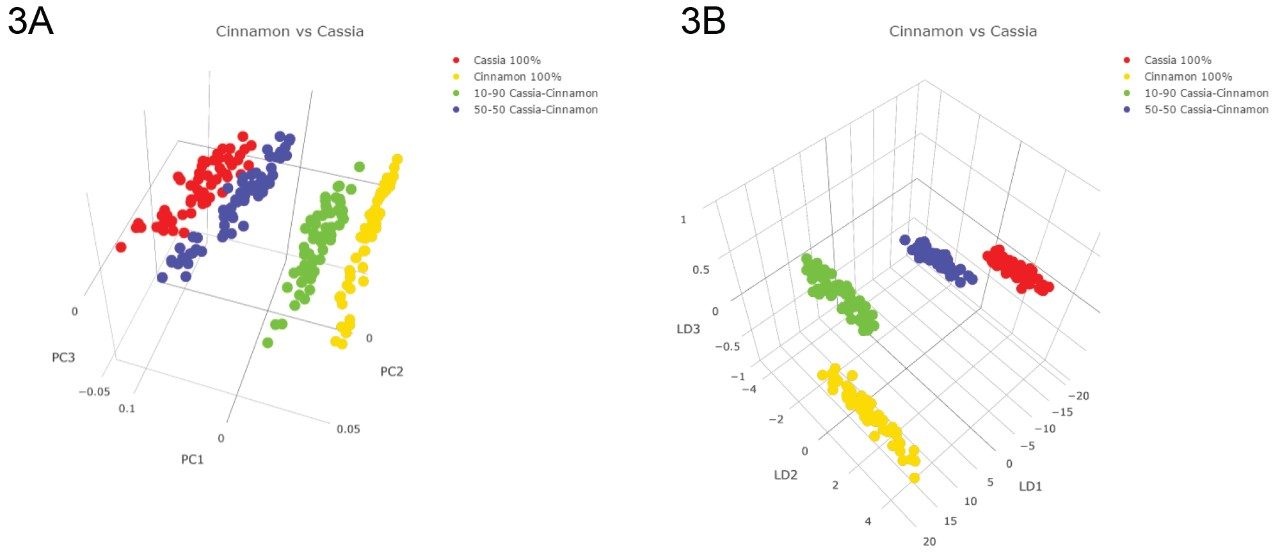
All chemometric models were calculated using the region of 100 to 300 m/z, as no significant features were observed above 300 m/z. For model training purposes, 10 PCA components and two LDA components were used. Class related clustering was apparent within the three-dimensional (3D) PCA scores plot using components 1, 2, and 3 (Figure. 3A). The combination of PCA (for data dimension reduction) and the supervised LDA generated four discrete class groupings within a five standard deviation outlier threshold are shown in Figure 3B.
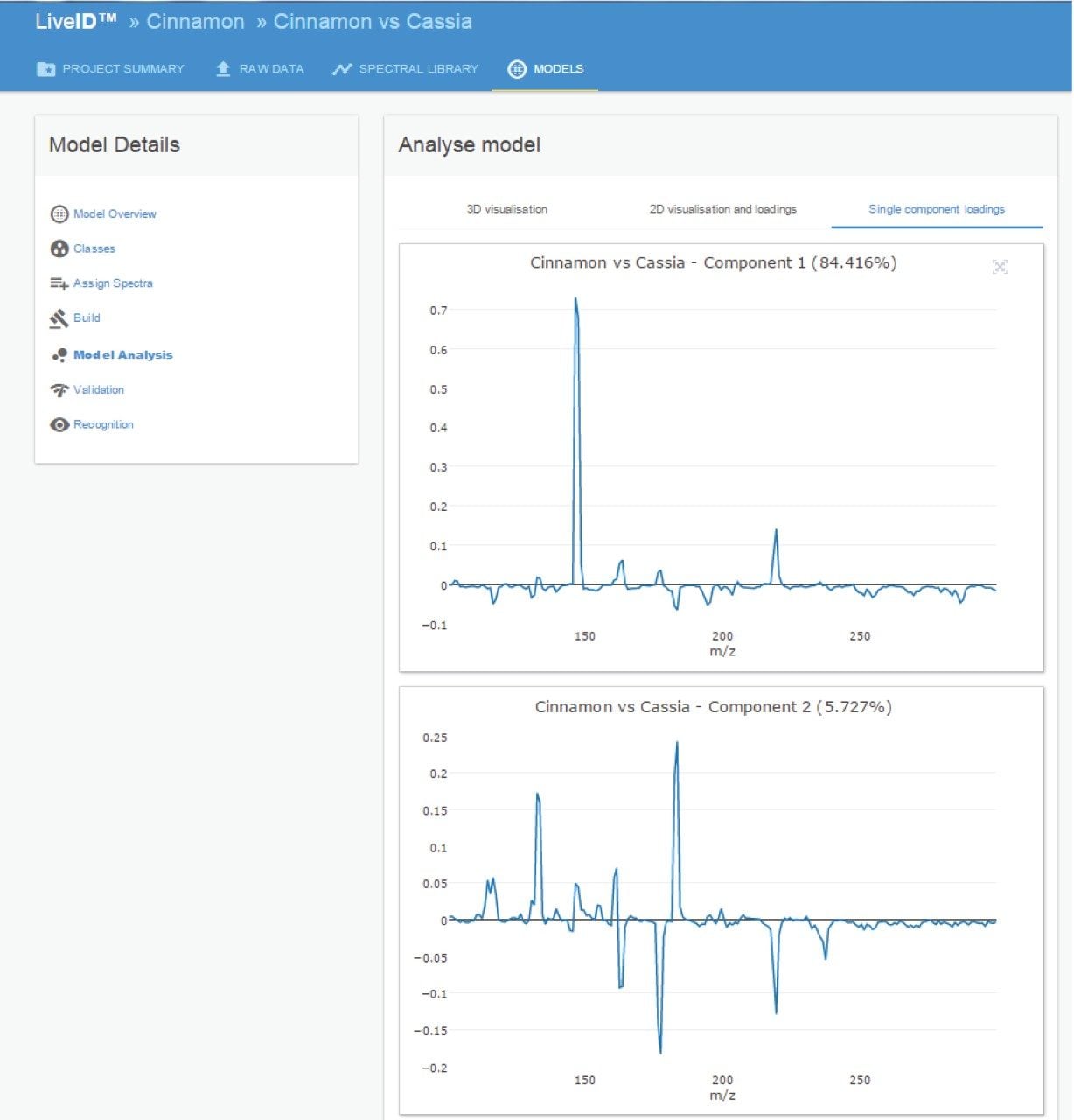
The loadings plot (Figure 4) shows the significant ions contributing to the class level discrimination. The ion at 147 m/z (coumarin) is seen to be the major feature in PC 1 accounting for c. 84% of the variance. The ions at 133 (cinnamaldehyde), 163 (methyl cinnamate), 177 (cinnamyl acetate), and 183 m/z (unknown) are contributory features in PC 2 accounting for c. 6% of the variance.
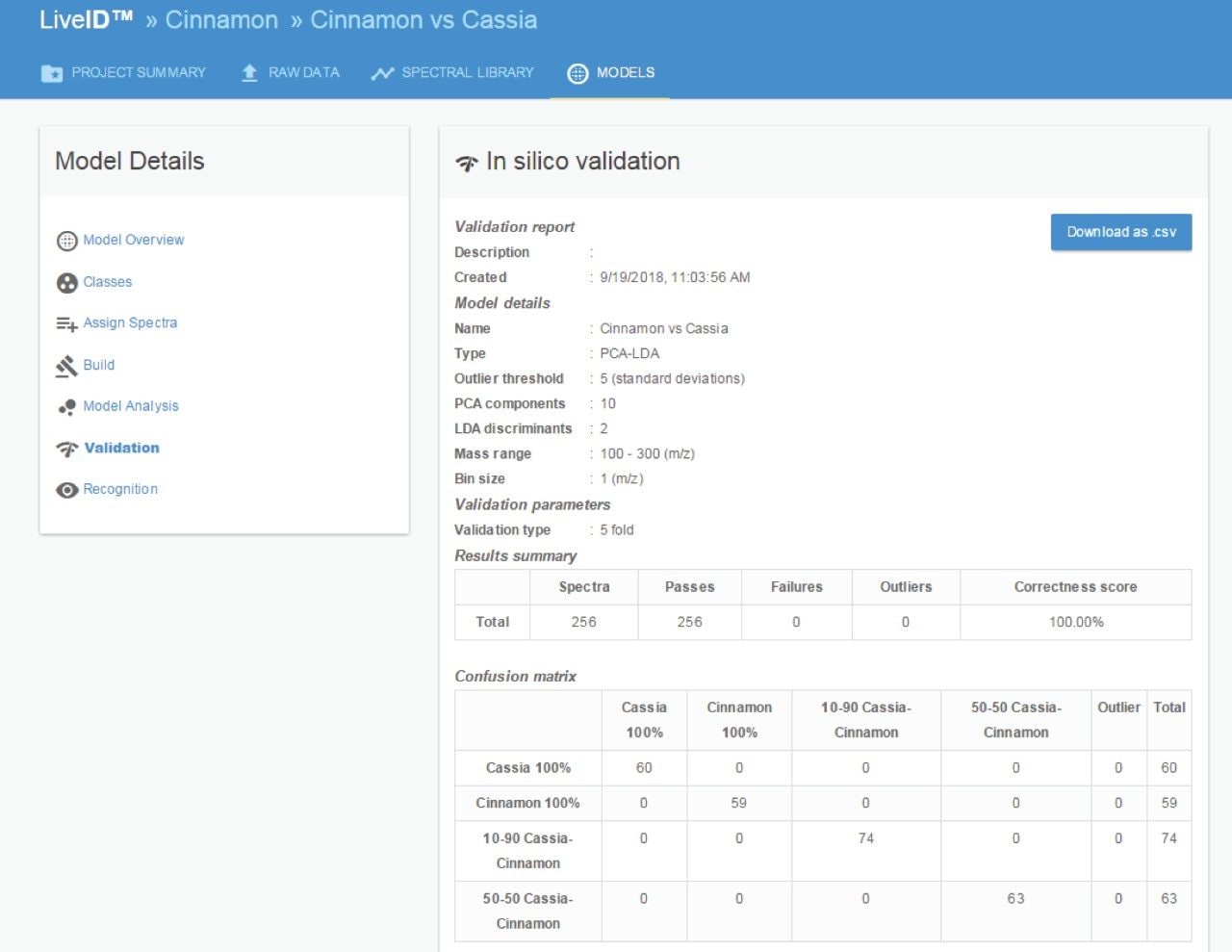
The cinnamon authenticity PCA/LDA model was subject to in silico cross validation using the “leave 20% out” method (Figure 5). The validation resulted in a 100% correct classification with no missed classifications and no outliers. The PCA/LDA model was also validated according to the “leave one file out” method, whereby each of the training data files was systematically excluded from the model and classified as an independent sample – which also resulted in a 100% correct classification rate.
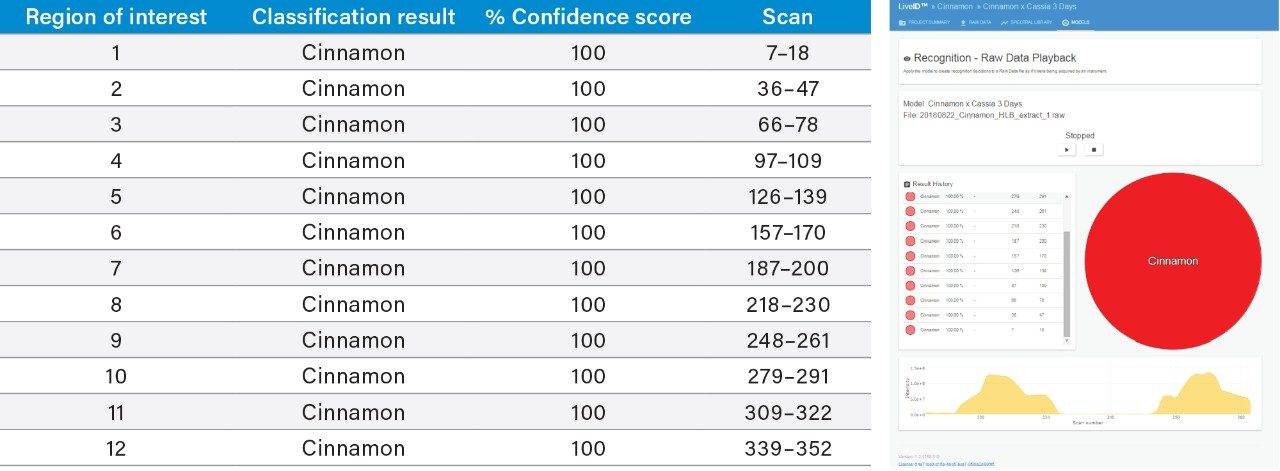
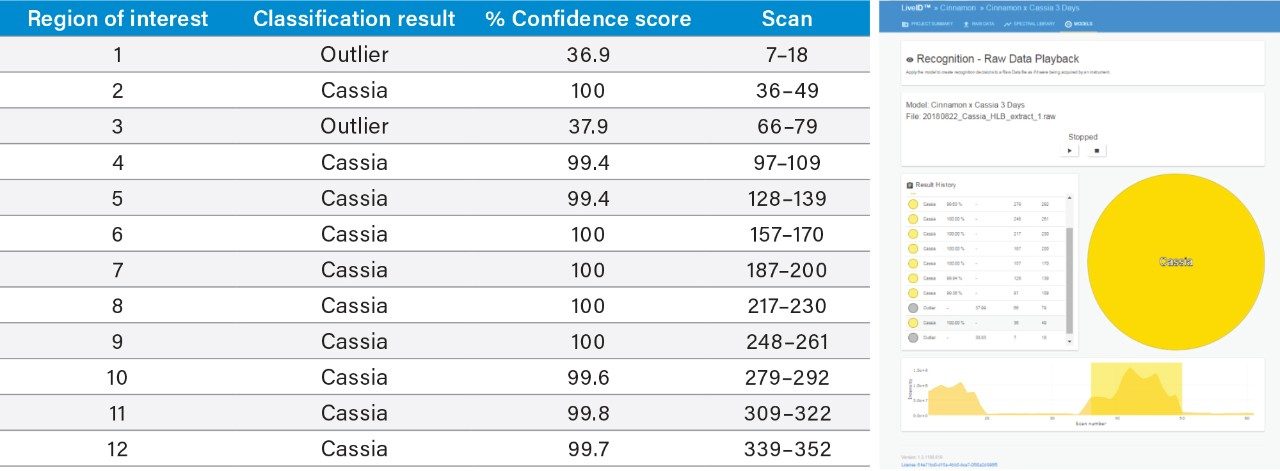
An inter-laboratory validation experiment was performed whereby new extracts of the authentic C. verum and C. cassia samples used for model training were prepared and analyzed in a second laboratory (Queen’s University Belfast) using a different DART QDa LiveID System. The mass spectral data (n=12 per sample) obtained was then classified using the LiveID model in playback recognition mode and the results were recorded (Tables 1A and 1B). All 12 replicate measurements of C. verum sample were accurately classified with high confidence scores, and 10 out of the 12 replicate measurements of C. cassia sample were accurately classified with two replicates being reported as “outliers”.
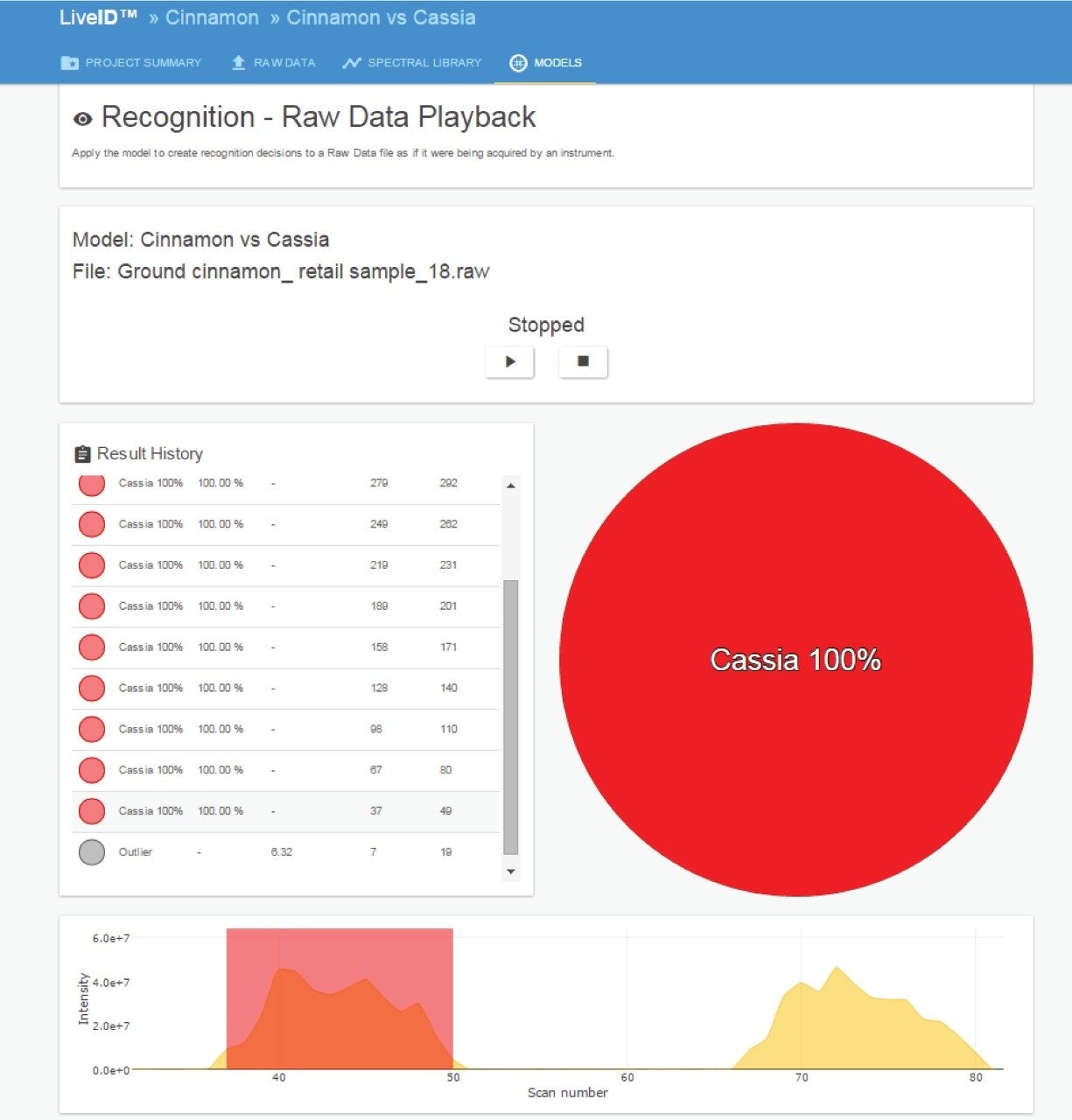
As a further test of model robustness, a sample of ground spice labelled as Cinnamon, with ingredients declared as C. cassia, was purchased from a local retail outlet and analyzed using the described cinnamon authenticity method. The extract was analyzed using LiveID in real-time recognition mode. QuickStrip position 1 was used as a reagent blank and the results from 11 replicate measurements, shown in Figure 6, classified the retail sample as 100% C. cassia with a high confidence score.
The authors kindly acknowledge John Hill, British Spice and Pepper Board, for the provision of the authentic Cinnamomum species samples.
720006409, October 2018