Automated ProteinWorks™ Sample Preparation Using Andrew+™ Pipetting Robot for Gelatin Speciation Analysis with ACQUITY™ Premier UPLC™ and Xevo™ TQ Absolute
Abstract
Gelatin is a popular ingredient in foods and pharmaceutical products due to its unique gelling properties. However, gelatin from a porcine source is considered a non-halal ingredient and cannot be present in products for the consumption by the Muslim community. Thus, gelatin speciation analysis is important to determine the gelatin source. In our previous work, a gelatin speciation analysis workflow was established using the ProteinWorks kit for protein digestion. The 3-step ProteinWorks protocol involves multiple repetitive reagent addition steps with a lot of manual pipetting, making it ideal for automation. In this work, a modified ProteinWorks 3-step protocol was automated using the Andrew+ pipetting robot to further increase the efficiency of the sample preparation process. Determination of the peptide markers was carried out using LC-MS/MS (ACQUITY™ Premier UPLC coupled to a Xevo TQ Absolute Tandem Quadrupole Mass Spectrometer). The performance of the revised method was shown to be sensitive (LOD and LOQ for porcine markers at 0.01% and 0.025%), repeatable (% RSD < 8%) and reproduceable.
Benefits
- Maximizing productivity and efficiency with automated protocol using Andrew+ pipetting robot
- Minimized error in sample preparation with automated protocol that has less human intervention
- Maximum peptide detection sensitivity with ACQUITY Premier UPLC solution and Xevo TQ-Absolute
Introduction
Gelatin is a popular ingredient used in a variety of food and pharmaceutical products where the main source of gelatin manufacturing comes from either bovine or porcine origin.1 The species of animals used to prepare gelatin is often restricted and regulated for both health and religious grounds for example, gelatin from a porcine source is considered non-halal and is forbidden for consumption by the Muslim community.
In our previous work, we carried out speciation analysis for gelatin with a simple 3-step sample digestion protocol using the Auto-Express ProteinWorks kit.2 The tryptic peptides obtained were then analyzed by LC-MS/MS. Although easy-to-follow, the digestion protocol requires multiple repetitive reagent addition steps which can be laborious especially when the number of samples to be processed is large.
In this application note, the productivity and efficiency of the sample preparation is further improved by automating the modified 3-step ProteinWorks digestion protocol with the Andrew+ pipetting robot. With Peltier+, Andrew+ can quickly and actively control the heating and cooling of accessories. This is important as the 3-step digestion protocol requires two different temperatures for the denaturation and digestion steps. The ability to rapidly automate the heating and cooling process for the samples further minimizes human intervention as well as saving overall processing time.
The protocol is designed and executed using the OneLab software, a cloud-native software. It has an intuitive interface which allows fast learning even with no programming knowledge. OneLab software has a variety of parameter settings which allows users to customize protocols with flexibility according to the characteristics of the liquids to be handled. OneLab software platform also provides traceability where the steps of the protocol are recorded as scripts. An experimental report will be generated after every protocol execution, giving details such as the settings used for the steps, sample information and timestamp of execution for easy housekeeping.
In addition, the method was transferred to a system comprising ACQUITY Premier UPLC and Xevo TQ Absolute tandem quadrupole mass spectrometer, to maximize the sensitivity for detection of the bovine and porcine peptide markers. The performance of the revised method was established for the analysis of halal-labeled candy.
Experimental
Commercially available porcine gelatin reference standard (Sigma Aldrich) and ten different food samples containing gelatin were pre-treated with 50 mM ammonium bicarbonate. This was followed by heating of the standard at 80 oC to obtain the 15 mg/mL standard solution. Porcine gelatin standard was added prior to digestion to portions of porcine-free halal-labeled candy to provide a procedural calibration series from 0.01%–1%.
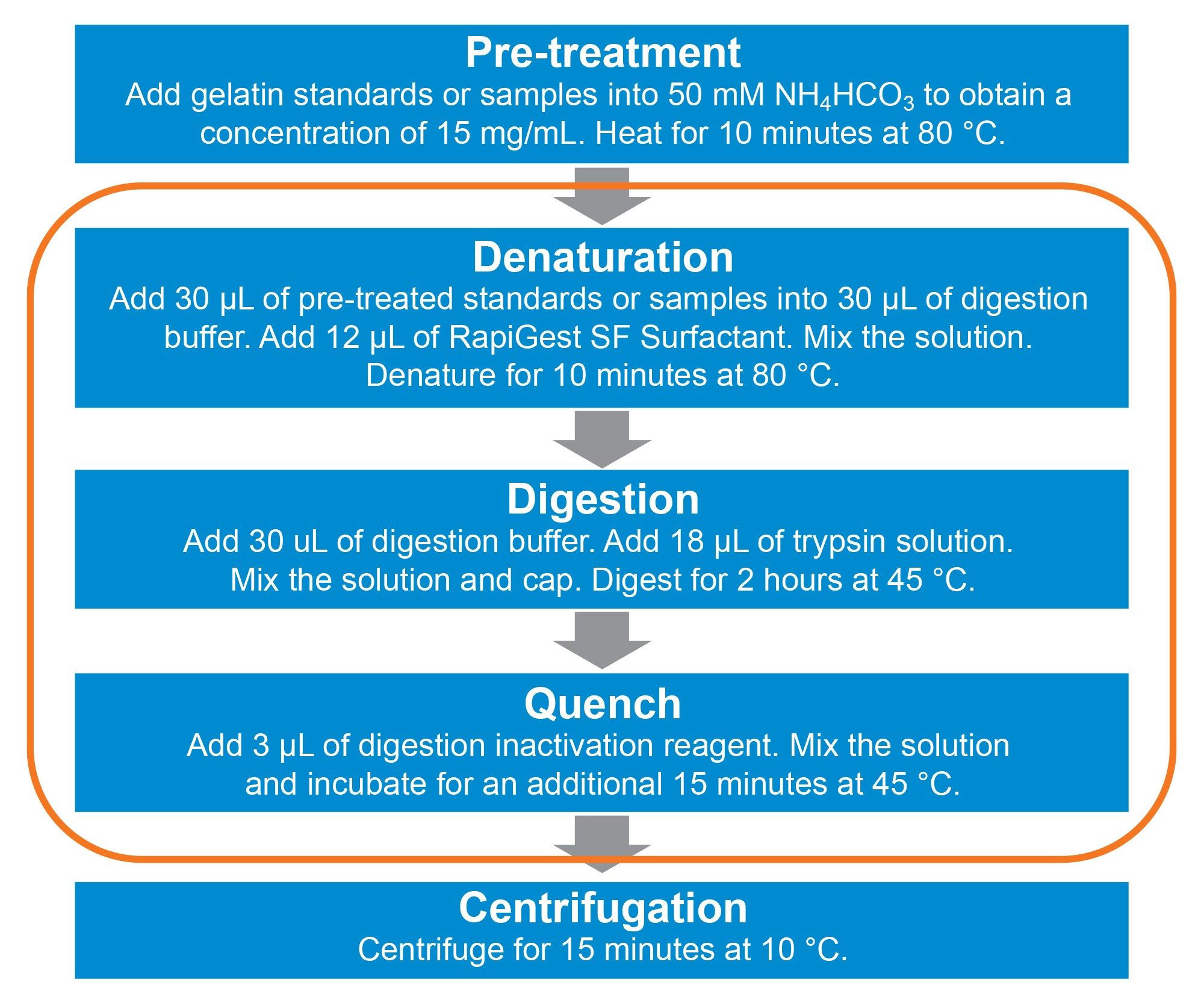
Andrew+ Pipetting Robot was used to automate the modified 3-step protocol using ProteinWorks Auto-eXpress Low 3 Digest Kit (p/n: 176004077) as shown in Figure 1. The protocol was designed and executed using OneLab™, a cloud-native software.
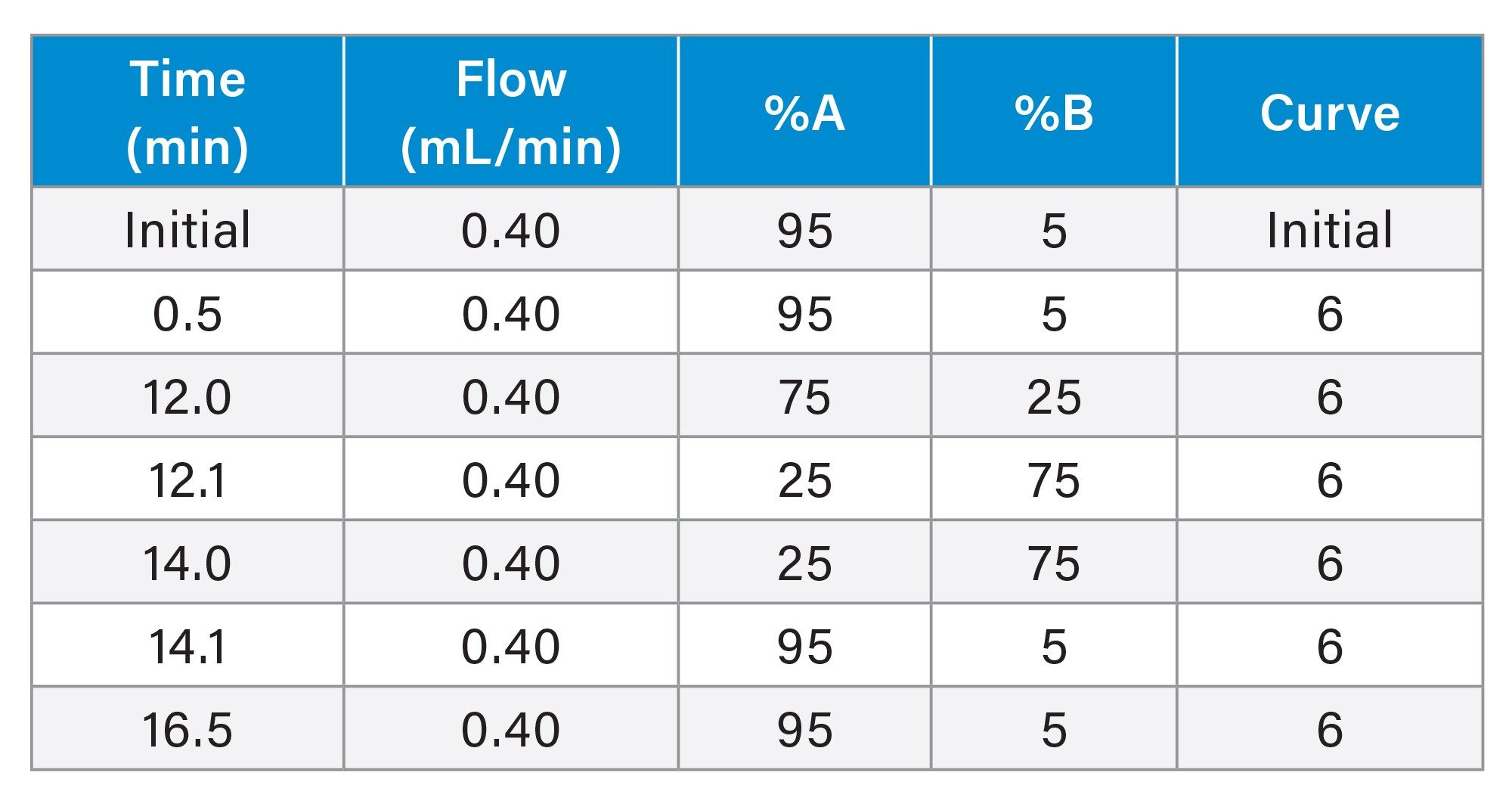
LC Conditions
|
LC system: |
ACQUITY Premier UPLC |
|
Column(s): |
ACQUITY Premier UPLC HSS T3 Column, 1.8 µm, 2.1 mm X 100 mm (p/n: 186009468) |
|
Column temperature: |
40 °C |
|
Injection volume: |
2 µL |
|
Flow rate: |
0.40 mL/min |
|
Mobile phase A: |
0.1% formic acid in water |
|
Mobile phase B: |
0.1% formic acid in acetonitrile |
Gradient Table
MS Conditions
|
MS system: |
Xevo TQ Absolute |
|
Ionization mode: |
ESI+ |
|
Capillary voltage: |
1 kV |
|
Source temperature: |
130 °C |
|
Desolvation temperature: |
600 °C |
|
Cone gas flow: |
150 L/Hr |
|
Desolvation gas flow: |
1000 L/Hr |
|
Nebulizser gas flow: |
7.0 bar |
Data Management
|
Chromatography software: |
MassLynx™ v4.2 |
|
MS software: |
MassLynx v4.2 |
|
Informatics: |
TargetLynx™ v4.2 |
MRM Transitions
The data was collected using the same MRM transitions listed in the previous study.2
Results and Discussion
Automated Sample Preparation
The calibration standards preparation and digestion protocol require a lot of repetitive pipetting for steps such as the addition of reagents and diluent. These steps are laborious and time consuming especially with large numbers of sampless. The automation with the Andrew+ pipetting robot frees up the lab analyst’s time from these manual pipetting steps for higher value tasks.

Figure 3 shows the comparison of time taken for Andrew+ and a typical lab analyst to complete the pipetting steps in the 3-step digestion protocol (without the heating and cooling steps). Andrew+ completes the pipetting task at a rate approximately two times faster than manual pipetting. Andrew+ can increase the efficiency of the workflow while also reducing potential human error.
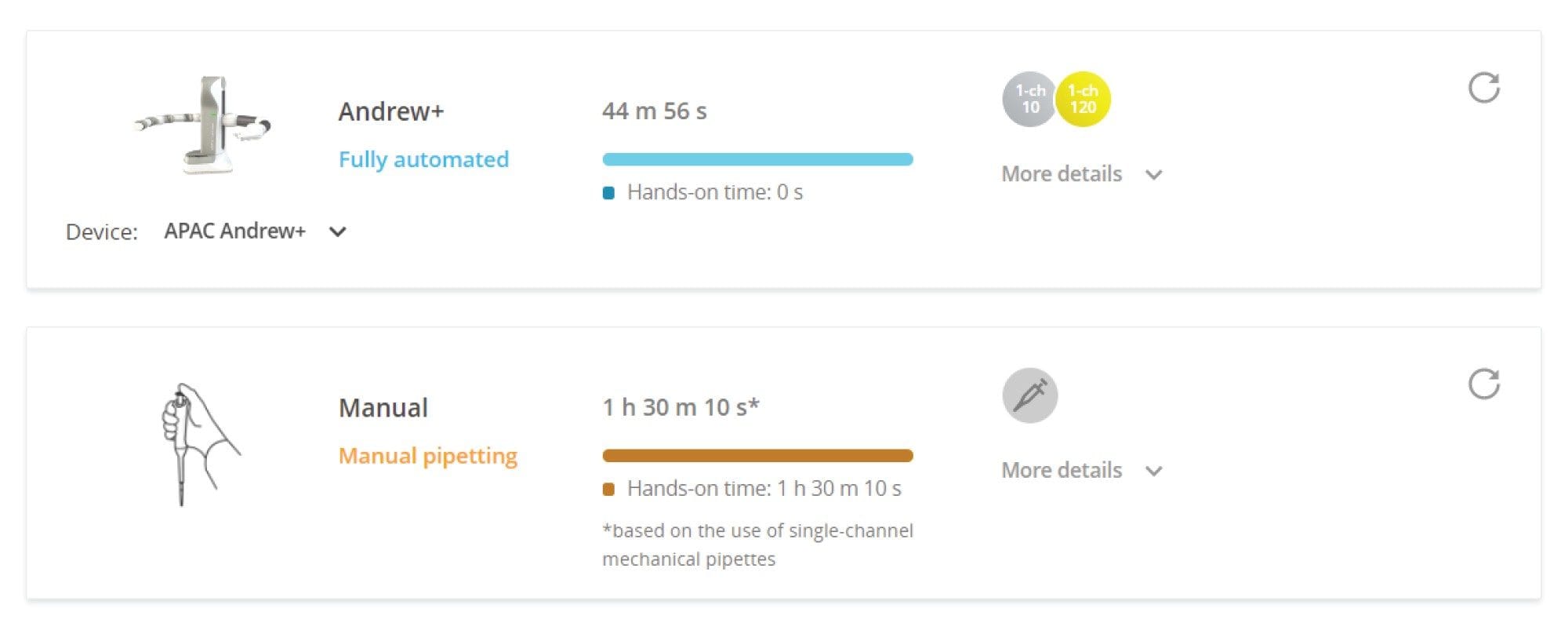
Heating and cooling steps needed in the 3-step digestion protocol can also be automated by Andrew+ with the addition of Peltier+, a connected device capable of rapid heating, and cooling of consumables. With a cooling rate of 9 oC/min, Peltier+ speeds up the cooling from 80 to 45 oC within minutes as compared to typical heating blocks which will take a much longer time to cool down passively. This feature also takes away the need to use two heating blocks to cater for two differing temperatures to save time.
The 3-step digestion protocol in this work has been modified from the previous study. The volume of trypsin used is reduced from 24 µL to 18 µL. The 25% trypsin volume reduction increases the number of samples that can be processed for the same amount of trypsin stock, thus reducing the cost of analysis per sample.
Method Performance
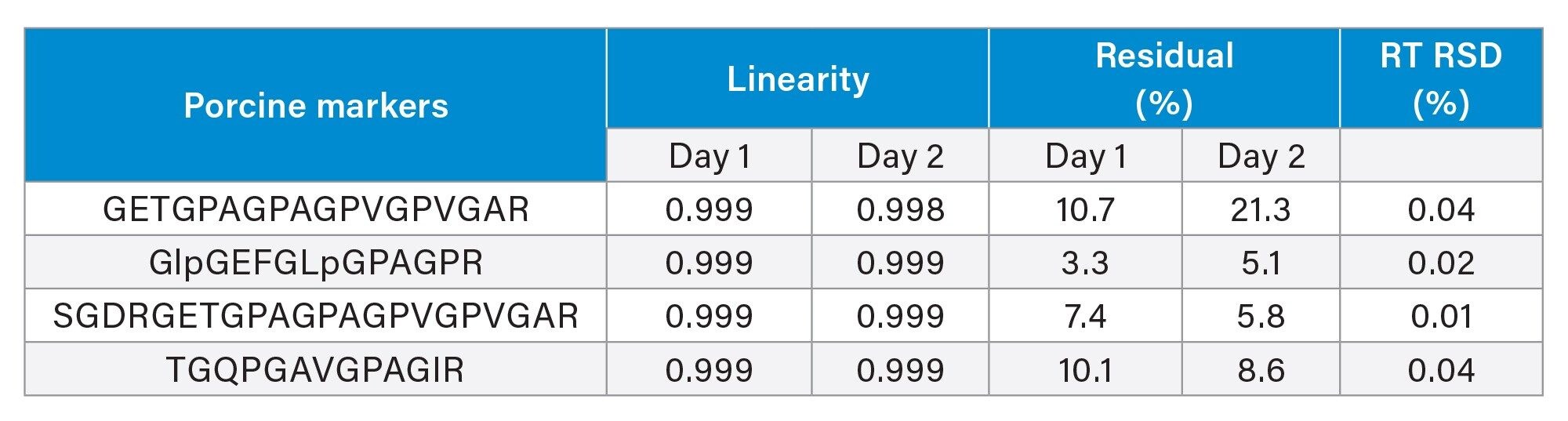
Procedural calibration curves were established for porcine gelatin in halal-labeled candy (0.025% to 1%). Two sets of procedural calibrants were prepared using Andrew+ on two separate days to evaluate the reproducibility when using the robot. Peaks were identified using retention time for the porcine markers which are stable, within 0.04% RSD.
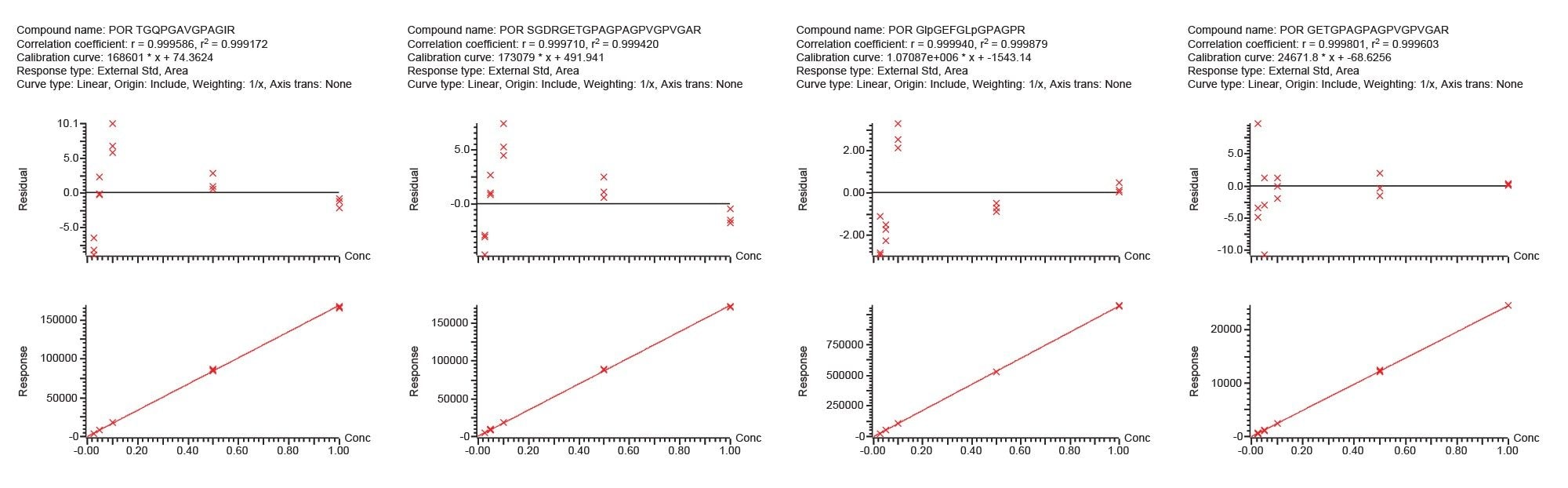
Figure 4 shows the calibration curve and percent residuals for the four porcine markers. The results obtained from both days gave comparable results, with coefficient of determination r2 >0.998 and residual percent <21% based on triplicate injections at each level (Table 1). The good linearity and accuracy show that the method is suitable for quantitative analysis.
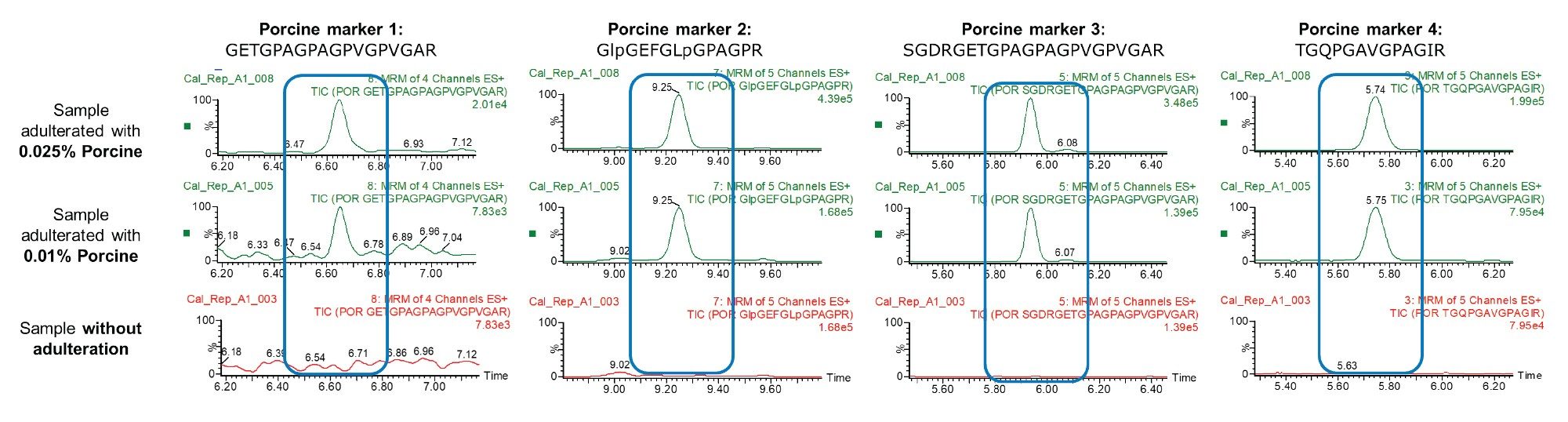
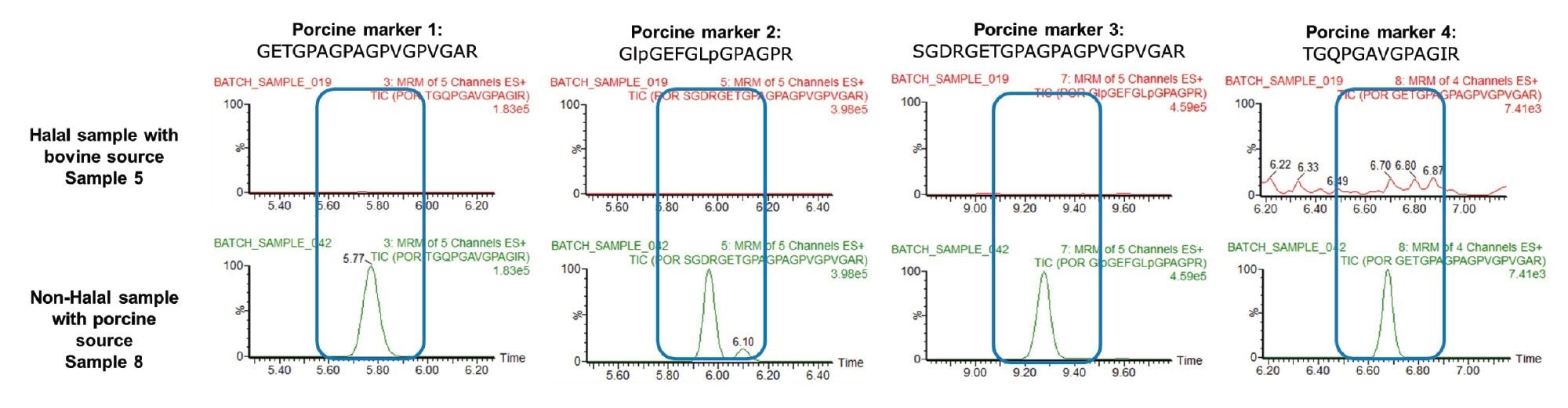
The sensitivity of the method was determined from evaluation of the signal/noise obtained from the chromatograms from analysis of spiked and blank candy. The limit of detection (LOD) and limit of quantitation (LOQ) were determined to be at 0.01% and 0.025 % porcine gelatin, respectively. Figure 5, shows the MRM chromatograms of halal-labeled candy matrix with and without porcine adulteration.
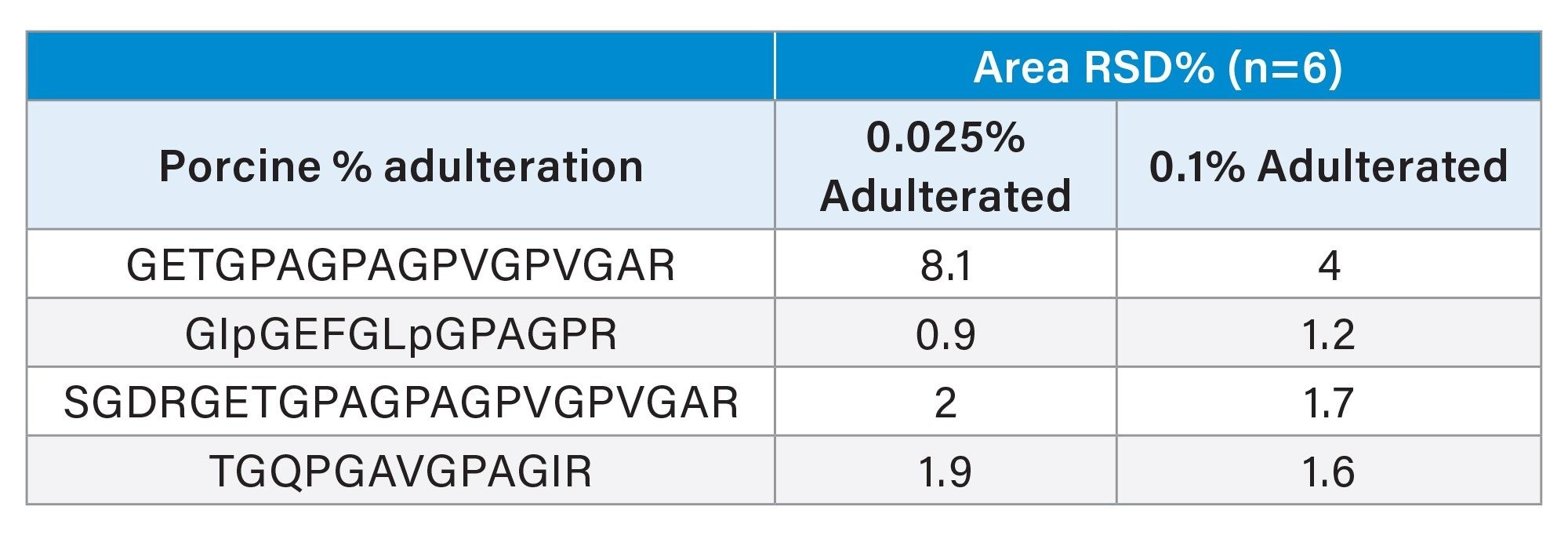
Repeatability was also evaluated by evaluation of the data from the replicate (n=6) analysis of halal-labeled candy spiked with porcine gelatin at 0.025% and 0.1%. The result shows good repeatability with RSDs for peak area > 8.1%.
Analysis of Candy Samples
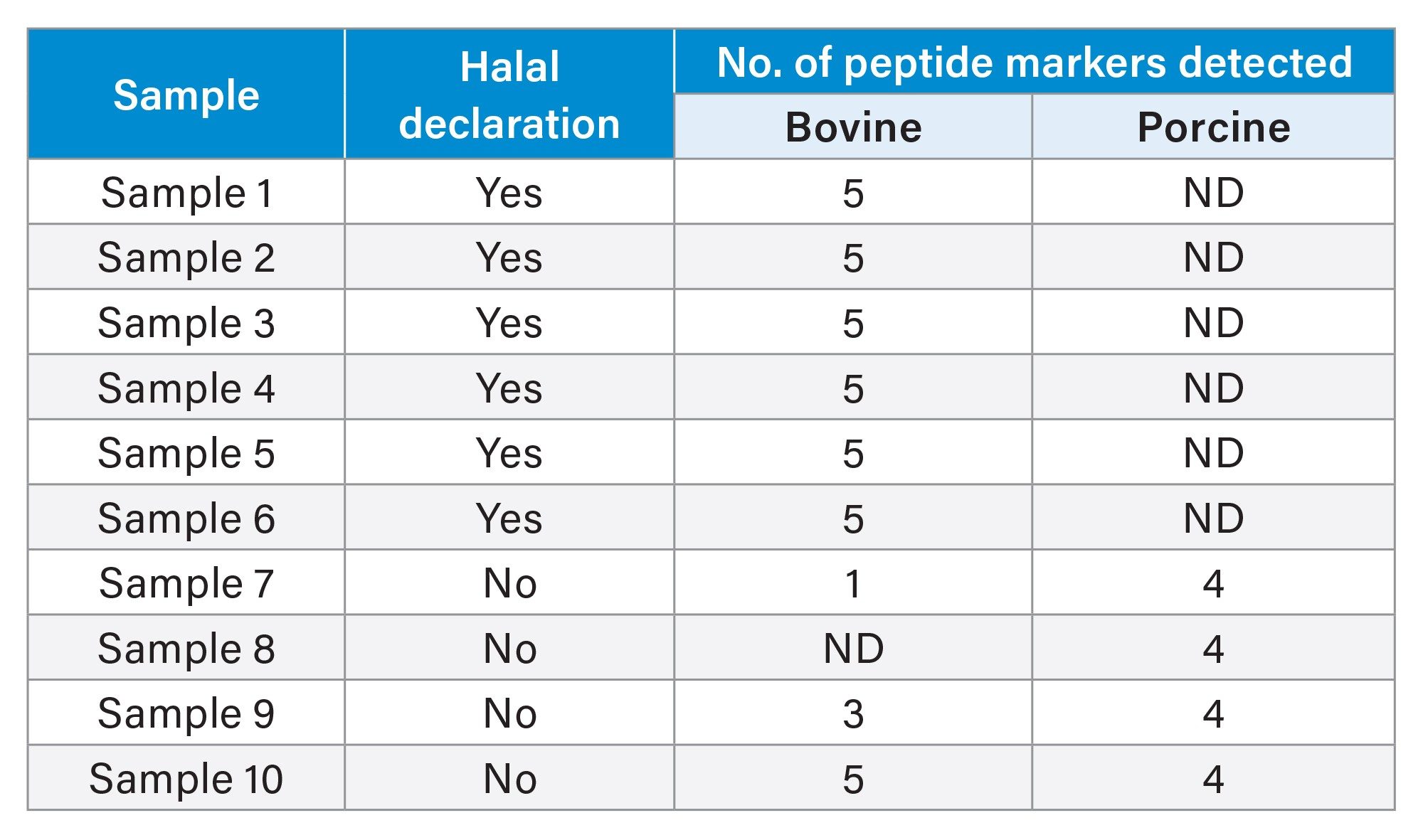
Ten commercial gelatin-containing candy samples were analyzed using the established method and the results are tabulated in Table 3. At least three of the MRM transitions with a minimum of signal-to-noise ratio of three must be present for a peptide marker to be detected and at least two peptide markers must be present for species assignment.3
Conclusion
This work demonstrates the automation of the sample preparation workflow with the ProteinWorks Auto-eXpress Low 3 Digest Kit using Andrew+ Pipetting Robot for gelatin speciation analysis. Modification of the trypsin volume used in the sample preparation workflow allows most cost-effective use of the ProteinWorks kit. The Andrew+ robot is able to complete the pipetting task at about two times 2x faster than manual pipetting. Using an LC-MS/MS system based upon the Xevo TQ Absolute, the method, with automation using Andrew+, was shown to be sensitive (LOD and LOQ for porcine at 0.01% and 0.025%), repeatable (% RSD < 8%) and reproduceable.
References
- Guo S, Xu X, Zhou X and Huang Y. A rapid and simple UPLC-MS/MS Method Using Collagen Marker Peptides for Identification or Porcine Gelatin. RSC Adv. 2018, 8: 3768–3773.
- Yin Ling Chew, Jun Xiang Lee, A Complete Solution for Gelatin Species Authentication in Routine Analysis Using ProteinWorks™ Auto-eXpress Digest Kit and Xevo™ TQ-XS. Waters Application Note, 720007667 2022.
- Song E, Gao Y, Wu C, Shi T, Nie S, Fillmore T L, Schepmoes A A, Gritsenko M A, Qian W, Smith R D, Rodland K D, Liu T. Targeted Proteomic Assays For Quantitation of Proteins Identified by Proteogenomic Analysis of Ovarian Cancer. Sci. Data (2017), 1–13.
720007894, May 2023