For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates to harmonize Skyline SRM/MRM software and Xevo TQ-S MS method development to enable the intuitive, prompt, and seamless development of multiple SRM/MRM peptide transitions and methods.
Move quickly from proteomics discovery to quantitative SRM/MRM-based validation with reliable, easy-to-use software for transition selection, automated MS-method building, and intuitive data.
Targeted LC-MS/MS SRM/MRM validation has the potential to bridge protein biomarker discovery and validation. Candidate proteins are typically identified in smaller scale relative quantification discovery studies. Typical bottlenecks observed are the selection of the appropriate proteotypic peptides for SRM/MRM quantitation and the development of the associated LC-MS methods for triple quadrupole mass spectrometers. Software plays a vital role in selecting the correct peptides to be measured, especially in the case of complex experiments, as a substitute for the target protein. This transitions into measurement of target peptides, where optimization for each peptide is typically performed. In this technology brief, we describe the use of the Skyline for peptide and transition selection process and method development for a Waters Xevo TQ-S Mass Spectrometer, including the results for the analysis allergenic peptide standards in a complex synthetic matrix. Skyline is an open source document editor for creating and analyzing targeted proteomics experiments.1
Five allergenic peptide standards were spiked in a matrix and the best candidate transitions predicted using Skyline, a software application for building SRM/MRM quantitative methods and analyzing the resulting mass spectrometer data. Skyline is vendor neutral and it was developed for creating and iteratively refining targeted methods for large-scale studies. A scheduled SRM/MRM method was created in order to independently optimize the collision energy for all peptides. The retention times were predicted with the built-in model of Skyline using the observed retention time as input/landmarks for linear retention time prediction and curve optimization/adjustment.
Figure 1 shows the predicted transitions for the allergenic protein of interest and the use of the Skyline Export Method editor for creating a retention time-scheduled Xevo TQ-S collision energy optimization experiment. A set of 11 collision energies was created, ranging from -5 V to +5 V, versus the optimally predicted collision energy derived from empirical observations for each peptide. The created method can be directly read and edited by the Experiment Setup SRM/MRM method editor of MassLynx Software, shown in Figure 2. The dwell times are automatically calculated based on the average chromatographic peak width. After the experiment is run, the results can either be reviewed in MassLynx or Skyline software. Figure 3 overlays the results of 11 SRM/MRM experiment for one of the peptides of interest. In this instance, the optimal collision energy was found to be 2 V lower than the predicted value and in agreement with the MassLynx Software data observed values, which were obtained via infusion of the peptide standards.
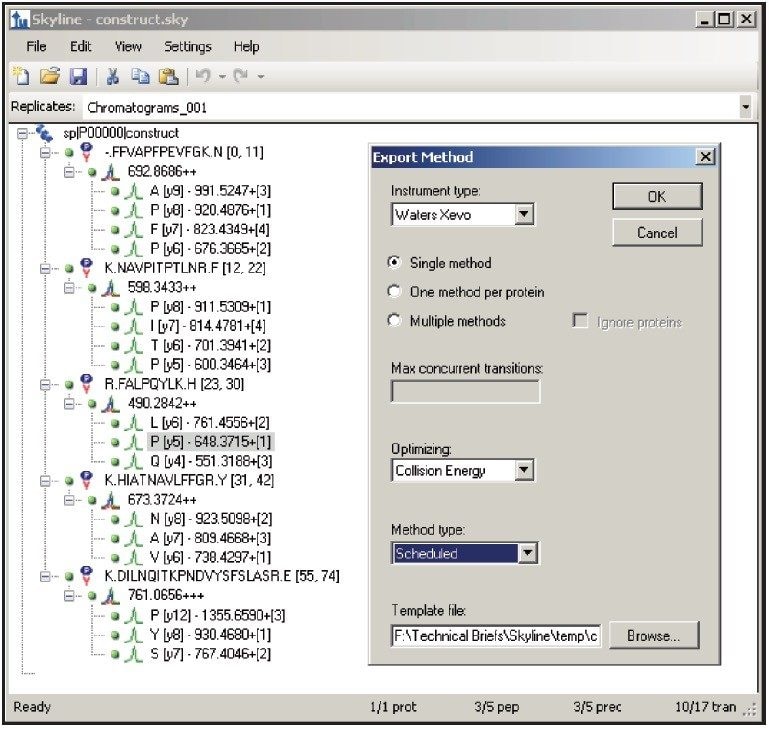
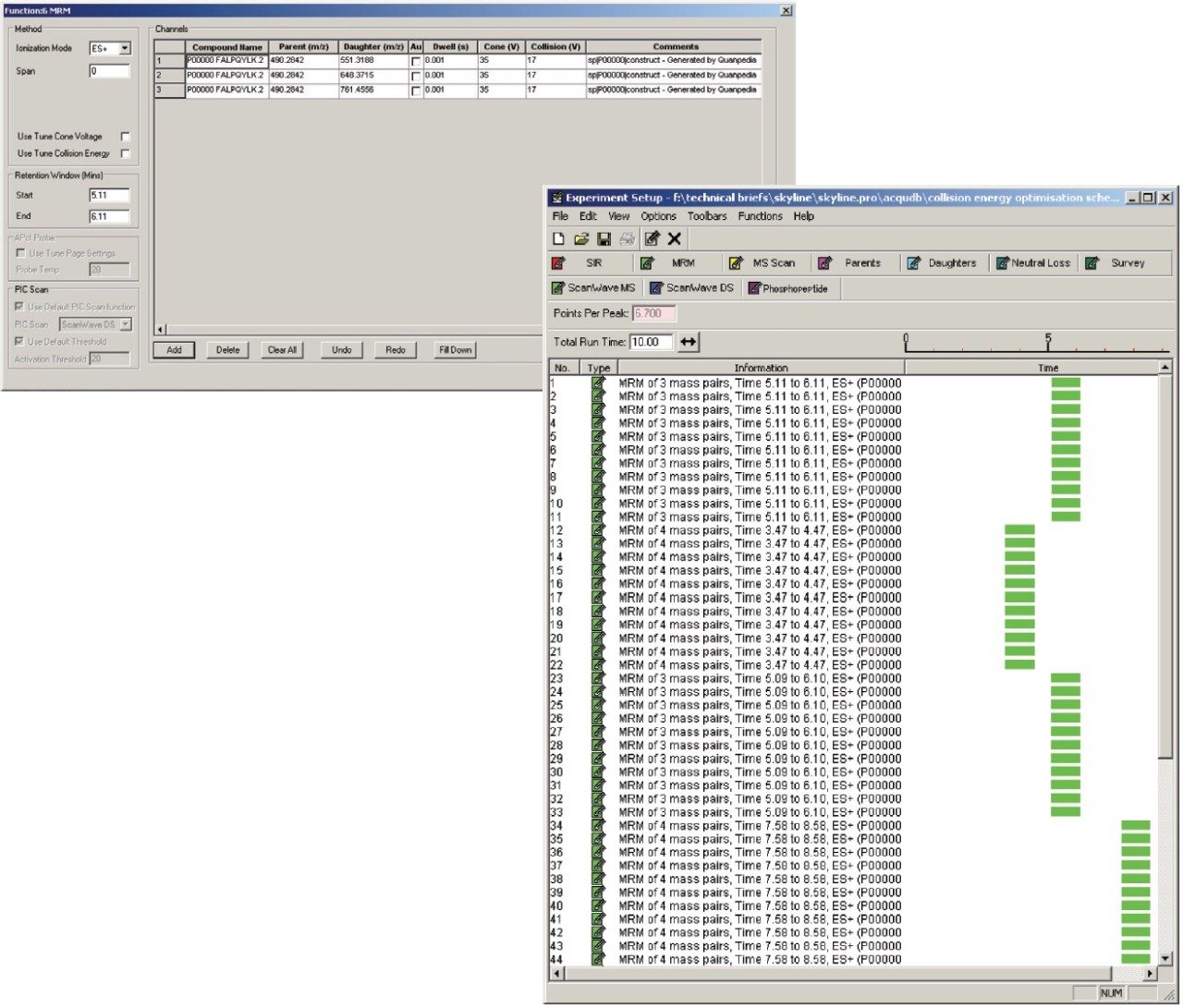
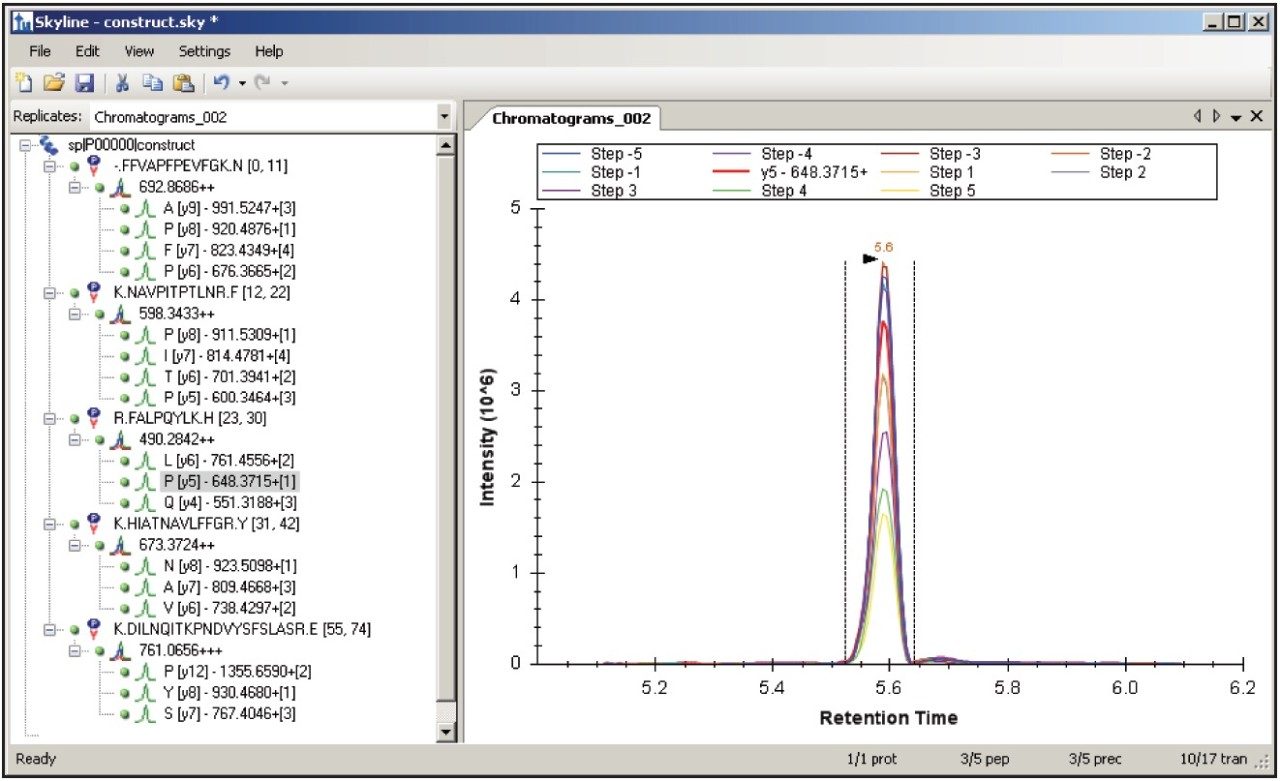
The SRM/MRM method development of five allergenic peptide standards spiked into a complex synthetic background for Xevo TQ-S has been demonstrated using Skyline opensource software.
720004346, May 2012