Conventional approaches to the preparation of N-glycans for HILIC-FLR-MS are either laborious, time-consuming, or require compromises in sensitivity. With the development of the GlycoWorks RapiFluor-MS N-Glycan Kit, we address these shortcomings by enabling unprecedented sensitivity for glycan detection while also improving the throughput of N-glycan sample preparation.
With this approach:
Glycoproteins are deglycosylated in approximately 10 minutes to produce N-glycosylamines.
These glycans are then rapidly reacted with the novel RapiFluor-MS reagent within a 5-minute reaction and are thereby labeled with a tag comprised of an efficient fluorophore and a highly basic tertiary amine that yields enhanced sensitivity for both fluorescence and MS detection.
In a final step requiring no more than 15 minutes, the resulting RapiFluor-MS labeled glycans are extracted from reaction byproducts by means of μElution HILIC SPE that has been rigorously developed to provide quantitative recovery of glycans (from neutral to tetrasialylated species) and to facilitate immediate analysis of samples.
Accordingly, an analyst can complete an N-glycan sample preparation, from glycoprotein to ready-to-analyze sample, in just 30 minutes when using the sensitivity enhancing RapiFluor-MS labeling reagent.
The N-glycan profile of a biopharmaceutical is commonly defined as a critical quality attribute, since it can be a measure of efficacy, safety, and manufacturing conditions.1-2 Therefore, it is important that approaches for the glycan analysis of clinical and commercial biotherapeutic formulations exhibit high sensitivity and facilitate detailed characterization. Additionally, it would be highly advantageous if such an analysis could also be performed with rapid turnaround times and high throughput capacity to expedite product development. Most analytical strategies for evaluating N-glycans from glycoproteins involve deglycosylation via PNGase F and the labeling of the resulting N-glycans with a chemical moiety that imparts a detectable attribute. In one, highly effective approach, labeled glycans are separated by hydrophilic interaction chromatography (HILIC) and detected by fluorescence (FLR) and sometimes mass spectrometry (MS).3-10
Unfortunately, conventional approaches to the preparation of N-glycans for HILIC-FLR-MS are either laborious, time-consuming, or require compromises in sensitivity.11 For instance, a conventional deglycosylation procedure requires that a glycoprotein sample be incubated for about 1 hour, while many analysts generically employ an overnight (16 hour) incubation. Combined with this process is a lengthy, 2 to 3 hour labeling step that relies on reductive amination of reducing, aldehyde termini that form on N-glycans only after they hydrolyze from their glycosylamine forms. And in the case of one of the most frequently employed labeling compounds, 2-aminobenzamide (2-AB), the resulting glycans can be readily detected by fluorescence but are rather challenging to detect by electrospray ionization mass spectrometry (ESI-MS).
Variations to conventional approaches for N-glycan sample preparation have been explored, but have not, as of yet, presented a solution that combines the desired attributes of simplicity, high MS sensitivity, and high throughput. Alternative labeling reagents, for example procainamide, that have functional groups to enhance electrospray ionization efficiency have been used,12 but this does not address the cumbersome, time consuming nature of relying on a reductive amination labeling step. Rapid tagging procedures that yield labeled glycans in a matter of minutes have consequently been investigated. In fact, two rapid tagging glycan labels were recently introduced, including a rapid tagging analog of aminobenzamide (AB).13 In a rapid reaction, the precursor glycosylamines of reducing, aldehyde terminated glycans are modified via a urea linked aminobenzamide. Although such a rapid tagging reagent accelerates the labeling procedure, it does not provide the enhanced ionization efficiencies needed in modern N-glycan analyses.
To address the above shortcomings, we have developed a sample preparation solution that enables unprecedented FLR and MS sensitivity for glycan detection while also improving the throughput of N-glycan sample preparation. A novel labeling reagent has been synthesized that rapidly reacts with glycosylamines upon their release from glycoproteins. Within a 5 minute reaction, N-glycans are labeled with RapiFluor-MS, a reagent comprised of an N-hydroxysuccinimide (NHS) carbamate rapid tagging group, an efficient quinoline fluorophore, and a highly basic tertiary amine for enhancing ionization. To further accelerate the preparation of N-glycans, rapid tagging has been directly integrated with a Rapid PNGase F deglycosylation procedure involving RapiGest SF surfactant and a HILIC µElution SPE clean-up step that provides highly quantitative recovery of the released and labeled glycans with the added benefit of not requiring a solvent dry-down step prior to the LC-FLR-MS analysis of samples.
|
LC system: |
ACQUITY UPLC H-Class Bio System |
|
Sample temp.: |
5 °C |
|
Analytical column temp.: |
60 °C |
|
Flow rate: |
0.4 mL/min |
|
Fluorescence detection: |
Ex 265/Em 425 nm (RapiFluor-MS) Ex 278/Em 344 nm (Instant AB) Ex 330/Em 420 nm (2-AB) (2 Hz scan rate [150 mm column]/5 Hz scan rate [50 mm column], Gain =1) |
|
Injection volume: |
≤1 µL (aqueous diluents with 2.1 mm I.D. columns) ≤30 µL (DMF/ACN diluted samples with 2.1 mm I.D. columns) |
|
Columns: |
ACQUITY UPLC Glycan BEH Amide 130Å, 1.7 µm, 2.1 x 50 mm ACQUITY UPLC Glycan BEH Amide 130Å, 1.7 µm, 2.1 x 150 mm |
|
Sample collection/ vials: |
Sample Collection Module Polypropylene 12 x 32 mm Screw Neck Vial, 300 µL volume |
|
Mobile phase A: |
50 mM ammonium formate, pH 4.4 (LC-MS grade; from a 100x concentrate) |
|
Mobile phase B: |
ACN (LC-MS grade) |
|
Time (min) |
Flow rate (mL/min) |
%A |
%B |
Curve |
|---|---|---|---|---|
|
0.0 |
0.4 |
25.0 |
75.0 |
6.0 |
|
11.7 |
0.4 |
46.0 |
54.0 |
6.0 |
|
12.2 |
0.2 |
100.0 |
0.0 |
6.0 |
|
13.2 |
0.2 |
100.0 |
0.0 |
6.0 |
|
14.4 |
0.2 |
25.0 |
75.0 |
6.0 |
|
15.9 |
0.4 |
25.0 |
75.0 |
6.0 |
|
18.3 |
0.4 |
25.0 |
75.0 |
6.0 |
|
Time (min) |
Flow rate (mL/min) |
%A |
%B |
Curve |
|---|---|---|---|---|
|
0.0 |
0.4 |
25.0 |
75.0 |
6.0 |
|
35.0 |
0.4 |
46.0 |
54.0 |
6.0 |
|
36.5 |
0.2 |
100.0 |
0.0 |
6.0 |
|
39.5 |
0.2 |
100.0 |
0.0 |
6.0 |
|
43.1 |
0.2 |
25.0 |
75.0 |
6.0 |
|
47.6 |
0.4 |
25.0 |
75.0 |
6.0 |
|
55.0 |
0.4 |
25.0 |
75.0 |
6.0 |
|
MS system: |
SYNAPT G2-S HDMS |
|
Ionization mode: |
ESI+ |
|
Analyzer mode: |
TOF MS, resolution mode (~20 K) |
|
Capillary voltage: |
3.0 kV |
|
Cone voltage: |
80 V |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
350 °C |
|
Desolvation gas flow: |
800 L/Hr |
|
Calibration: |
NaI, 1 µg/µL from 500–2500 m/z |
|
Lockspray (ASM B-side): |
100 fmol/µL Human Glufibrinopeptide B in 0.1% (v/v) formic acid, 70:30 water/acetonitrile every 90 seconds |
|
Acquisition: |
500–2500 m/z, 1 Hz scan rate |
|
Data management: |
MassLynx Software (v4.1) |
N-glycans from Intact mAb Mass Check Standard (p/n 186006552), bovine fetuin (Sigma F3004), and pooled human IgG (Sigma I4506) were prepared according to the guidelines provided in the GlycoWorks RapiFluor-MS N-Glycan Kit Care and Use Manual (715004793).
To compare the response factors of Instant AB and RapiFluor-MS labeled glycans, labeling reactions were performed with equivalent molar excesses of reagent, and crude reaction mixtures were directly analyzed by HILIC-FLR-MS in order to avoid potential biases from SPE clean-up procedures. Response factors were determined as ratios of the FA2 N-glycan (Oxford notation) chromatographic peak area to the mass of glycoprotein from which the glycan originated.
To compare the response factors of 2-AB labeled versus RapiFluor-MS labeled glycans, equivalent quantities of labeled N-glycans from pooled human IgG were analyzed by HILIC-FLR-MS. Column loads were calibrated using external quantitative standards of 2-AB labeled triacetyl chitotriose and RapiFluor-MS derivatized propylamine (obtained in high purity; confirmed by HPLC and 1H NMR). Response factors were determined as ratios of the FA2 chromatographic peak area to the mole quantity of glycan.
The procedure for extracting labeled RapiFluor-MS glycans after derivatization was evaluated using a test mixture containing N-glycans released and labeled from a 1:1 mixture (by weight) of pooled human IgG and bovine fetuin. The test mixture was prepared and then reconstituted in a solution equivalent in MS conditions MS system: SYNAPT G2-S HDMS Ionization mode: ESI+ Analyzer mode: TOF MS, resolution mode (~20 K) Capillary voltage: 3.0 kV Cone voltage: 80 V Source temp.: 120 °C Desolvation temp.: 350 °C Desolvation gas flow: 800 L/Hr Calibration: NaI, 1 µg/µL from 500–2500 m/z Lockspray (ASM B-side): 100 fmol/µL Human Glufibrinopeptide B in 0.1% (v/v) formic acid, 70:30 water/acetonitrile every 90 seconds Acquisition: 500–2500 m/z, 1 Hz scan rate Data management: MassLynx Software (V4.1) Rapid Preparation of Released N-Glycans for HILIC Analysis Using a Novel Fluorescence and MS-Active Labeling Reagent 4 composition to the solution glycans are subjected to when following the protocol of the RapiFluor-MS N-Glycan Kit. All other sample preparation techniques are described in the GlycoWorks RapiFluor-MS N-Glycan Kit Care and Use Manual (715004793).
A new labeling reagent for facilitating N-glycan analysis has been synthesized based on rational design considerations (Figure 1) that would afford rapid labeling kinetics, high fluorescence quantum yield, and significantly enhanced MS detectability. Conventional N-glycan sample preparation is dependent on reductive amination of aldehyde terminated saccharides, a process that requires glycans to undergo multiple chemical conversions and a lengthy high temperature incubation step.11 Moreover, glycans must be reductively aminated in anhydrous conditions in order to minimize desialylation. Sample preparations are therefore burdened with transitioning a sample from aqueous to anhydrous conditions. For these reasons, the newly designed labeling reagent foregoes reductive amination and instead takes advantage of an aqueous rapid tagging reaction. To this end, Waters has drawn upon its experience with rapid fluorescence labeling of amino acids to develop a new reagent that meets the needs of modern, N-glycan analysis. More than 20 years ago, Waters introduced a rapid tagging labeling reagent, known as AccQ•Fluor, that is now widely use to accurately profile the amino acid composition of protein samples via fluorescence detection.14-15
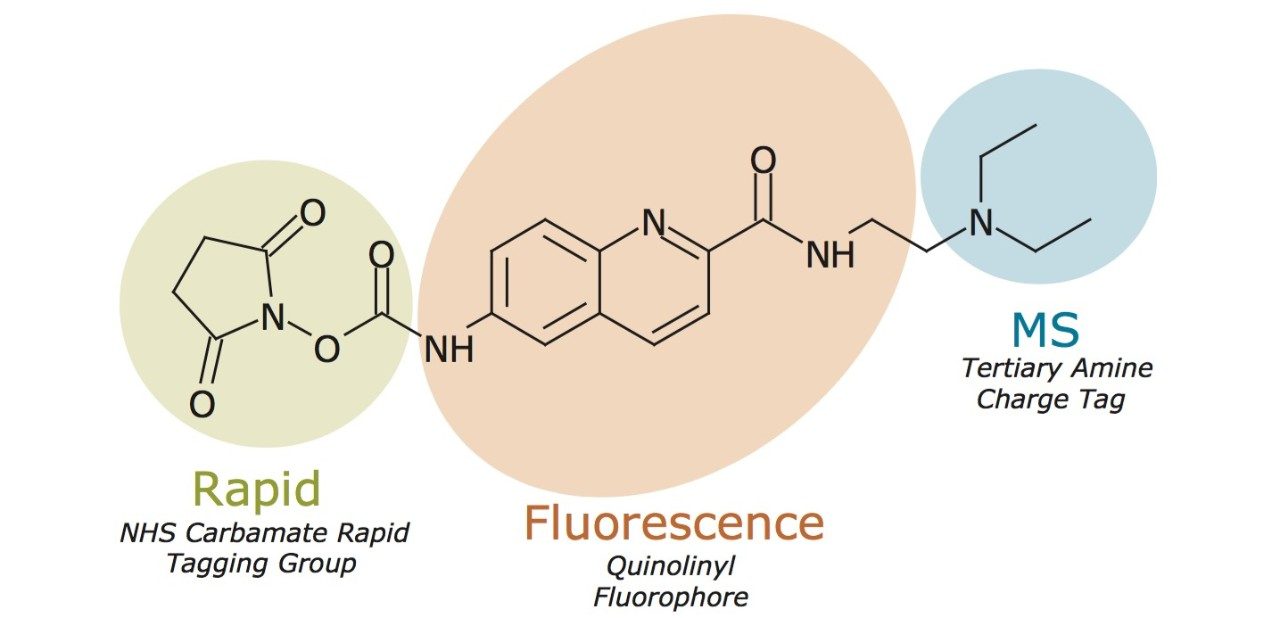
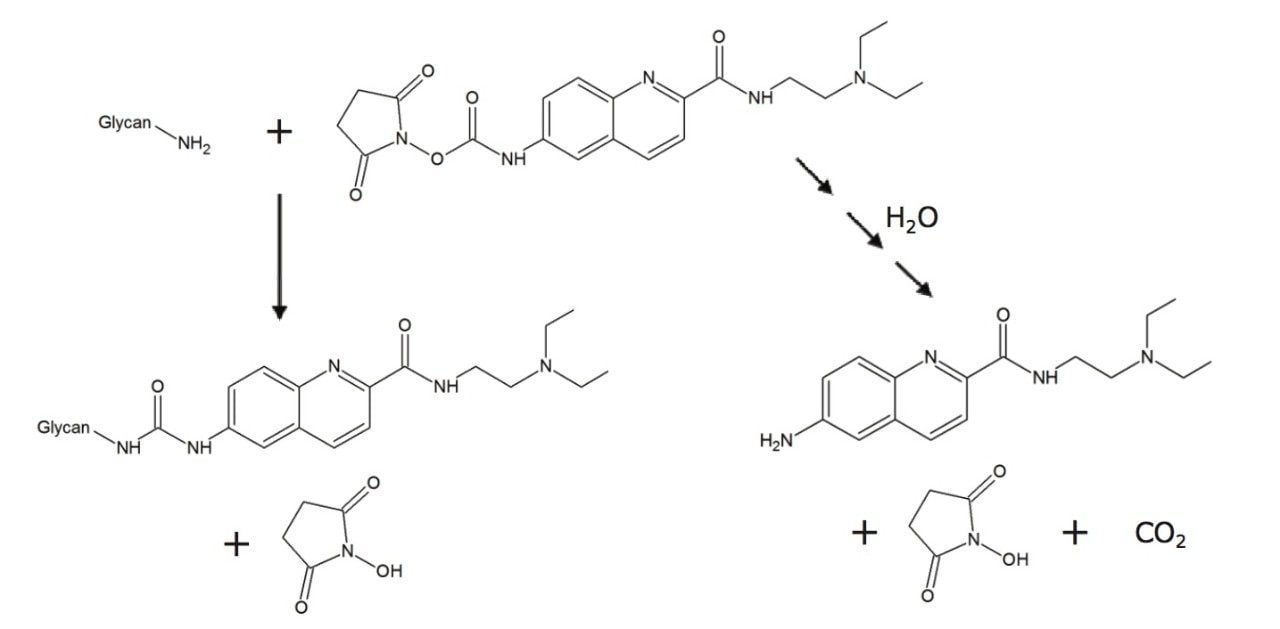
AccQ•Fluor possesses two important chemical characteristics: an NHS-carbamate rapid tagging reactive group and a highly efficient quinolinyl fluorophore. These features of AccQ•Fluor form the basis of the new glycan labeling reagent. The NHS-carbamate reactive group of this reagent enables glycosylamine bearing N-glycans to be rapidly labeled following their enzymatic release from glycoproteins. Within a 5 minute reaction, N-glycans are labeled with the new reagent under ambient, aqueous conditions to yield a highly stable urea linkage (Figure 2). In addition to rapid tagging capabilities, the new labeling reagent also supports high sensitivity for both MS and flourescence detection. A quinoline fluorophore serves as the central functionality of the new reagent that, as with AccQ•Fluor, facilitates high sensitivity fluorescence detection. In addition to AccQ•Fluor, however, the new reagent has been synthesized with a tertiary amine side chain as a means to enhance MS signal upon positive ion mode electrospray ionization (ESI+). In summary, the resulting N-glycan labeling reagent is built upon our expertise in chemical reagents and three important chemical attributes, a rapid tagging reactive group, an efficient fluorophore, and a highly basic MS active group. To describe these noteworthy attributes, the new labeling reagent has accordingly been named RapiFluor-MS.
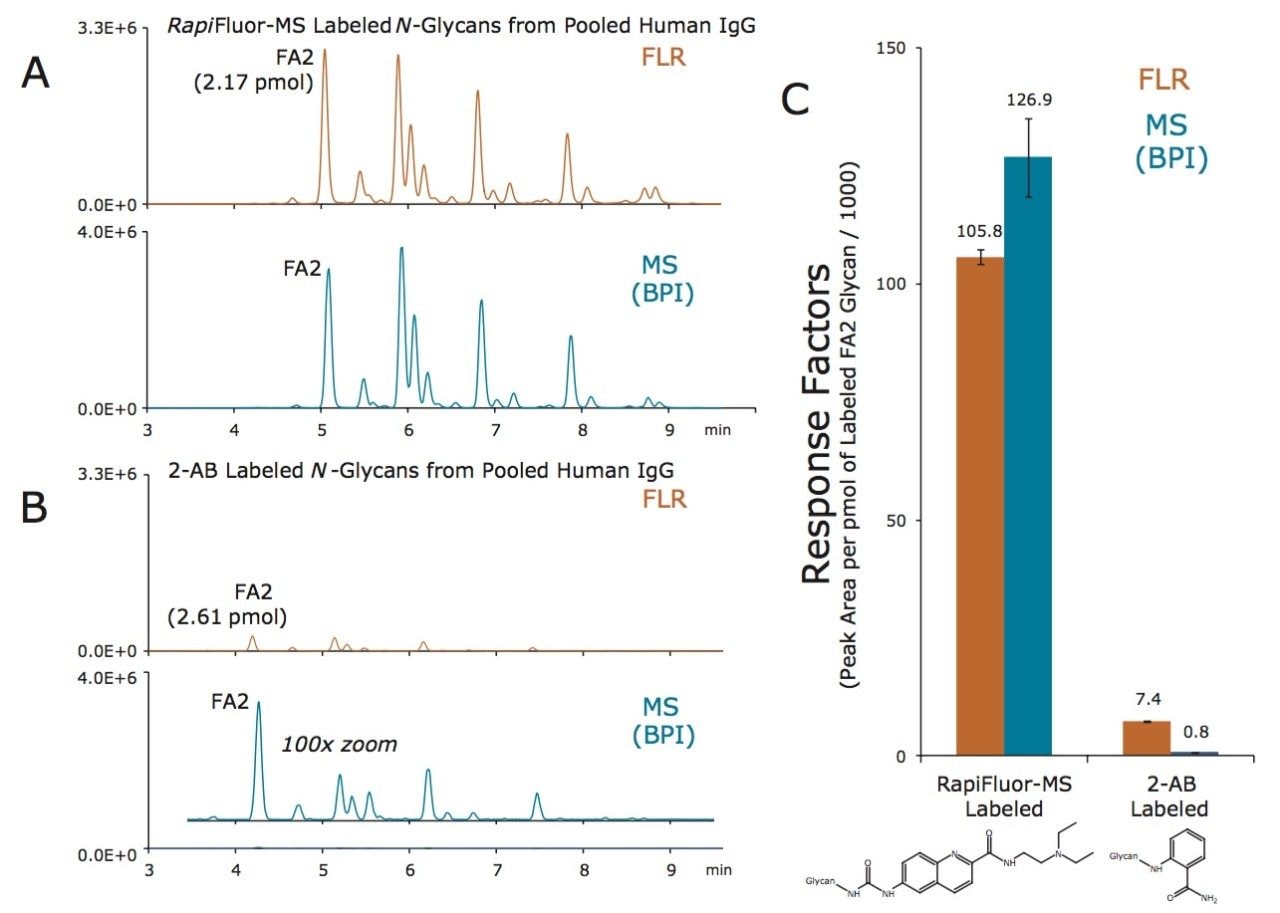
RapiFluor-MS N-glycan labeling has been extensively studied. In particular, the response factors of RapiFluor-MS labeled glycans have been benchmarked against those observed for glycans labeled with alternative reagents. The most closely related, commercially available alternative to RapiFluor-MS is an NHS carbamate analog of aminobenzamide, known as Instant AB.13 Figures 3A and 3B present HILIC fluorescence and base peak intensity (BPI) MS chromatograms for equivalent quantities of N-glycans released from a murine monoclonal antibody (Intact mAb Mass Check Standard, p/n 186006552) and labeled with RapiFluor-MS and Instant AB, respectively. Based on the observed chromatographic peak areas, response factors for fluorescence and MS detection were determined for the most abundant glycan in the IgG profile, the fucosylated, biantennary FA2 glycan (Figure 3C). Our results for the FA2 glycan indicate that RapiFluor-MS labeled glycans produce 2 times higher fluorescence signal and, more astoundingly, 780 times greater MS signal than N-glycans labeled with Instant AB.

In a similar fashion, RapiFluor-MS labeling has also been compared to conventional 2-AB labeling. To draw such a comparison, N-glycans released from pooled human IgG and labeled with either RapiFluor-MS or 2-AB were analyzed by HILIC-FLR-MS at equivalent mass loads (Figures 4A and 4B, respectively). Given that rapid tagging and reductive amination are performed by significantly different procedures, external calibrations were established using quantitative standards in order to determine the amounts of FA2 glycan loaded and eluted from the HILIC column. Response factors determined using these calibrated amounts of FA2 glycan are provided in Figure 4C. Again, it is found that RapiFluor-MS labeled glycans are detected with significantly higher signal (14 times higher fluorescence and 160 times greater MS signal versus 2-AB labeled glycans.)
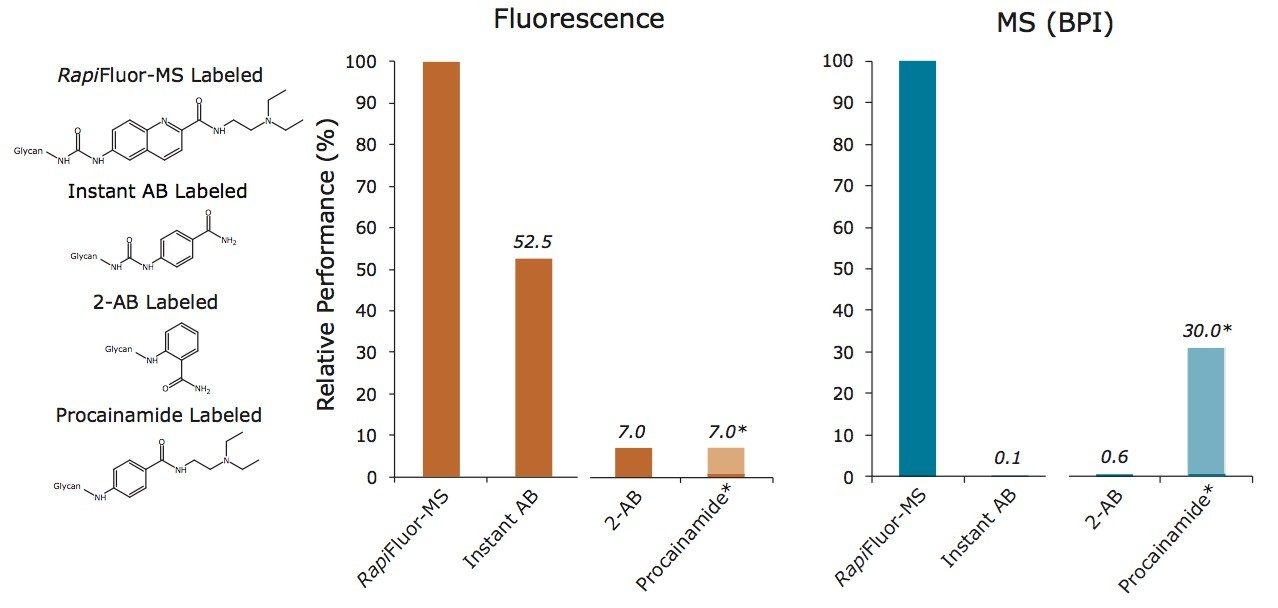
To summarize the above observations, we have plotted the response factors of Instant AB and 2-AB as percentages of the response factors of RapiFluor-MS (Figure 5). The gains in fluorescence and MS sensitivity are apparent in this plot, since it portrays response factors for Instant AB and 2-AB normalized to those for RapiFluor-MS. In this plot, the relative performance of reductive amination with another alternative labeling reagent, procainamide, is also provided. Procainamide is a chemical analog to aminobenzamide that has recently been exploited to enhance the ionization of reductively aminated glycans when they are analyzed by HILIC-ESI(+)-MS. Previous studies have shown that procainamide labeled glycans yield comparable fluorescence signal and up to 50 times greater MS signal when compared to 2-AB labeled glycans.12,16 Compared to procainamide, RapiFluor-MS will therefore provide sizeable gains in MS sensitivity. That is to say, the novel RapiFluor-MS labeling reagent not only supports rapid tagging of glycans but it also provides analysts with unmatched sensitivity for fluorescence and MS based detection.
RapiFluor-MS labeling revolutionizes N-glycan sample preparation and can be readily adopted in the laboratory with the GlycoWorks RapiFluor-MS N-Glycan Kit. This complete solution from Waters and New England BioLabs was purposefully designed to remove the bottlenecks from all aspects of N-glycan sample preparation. The optimized N-glycan sample preparation workflow requires a minimum of three steps, including deglycosylation (to release glycans from a glycoprotein), labeling (to impart a detectable chemical entity to glycans), and a clean-up step (to remove potential interferences from the sample) (Figure 6). Conventional approaches to N-glycan sample preparation can be very time consuming due to not only lengthy labeling procedures but also lengthy deglycosylation steps that range from 1 to 16 hours. To ensure rapid labeling with RapiFluor-MS was not encumbered by a time-consuming deglycosylation process, Waters partnered with New England BioLabs to co-develop a Rapid PNGase F deglycosylation procedure specifically designed for integration with rapid tagging labeling reagents.
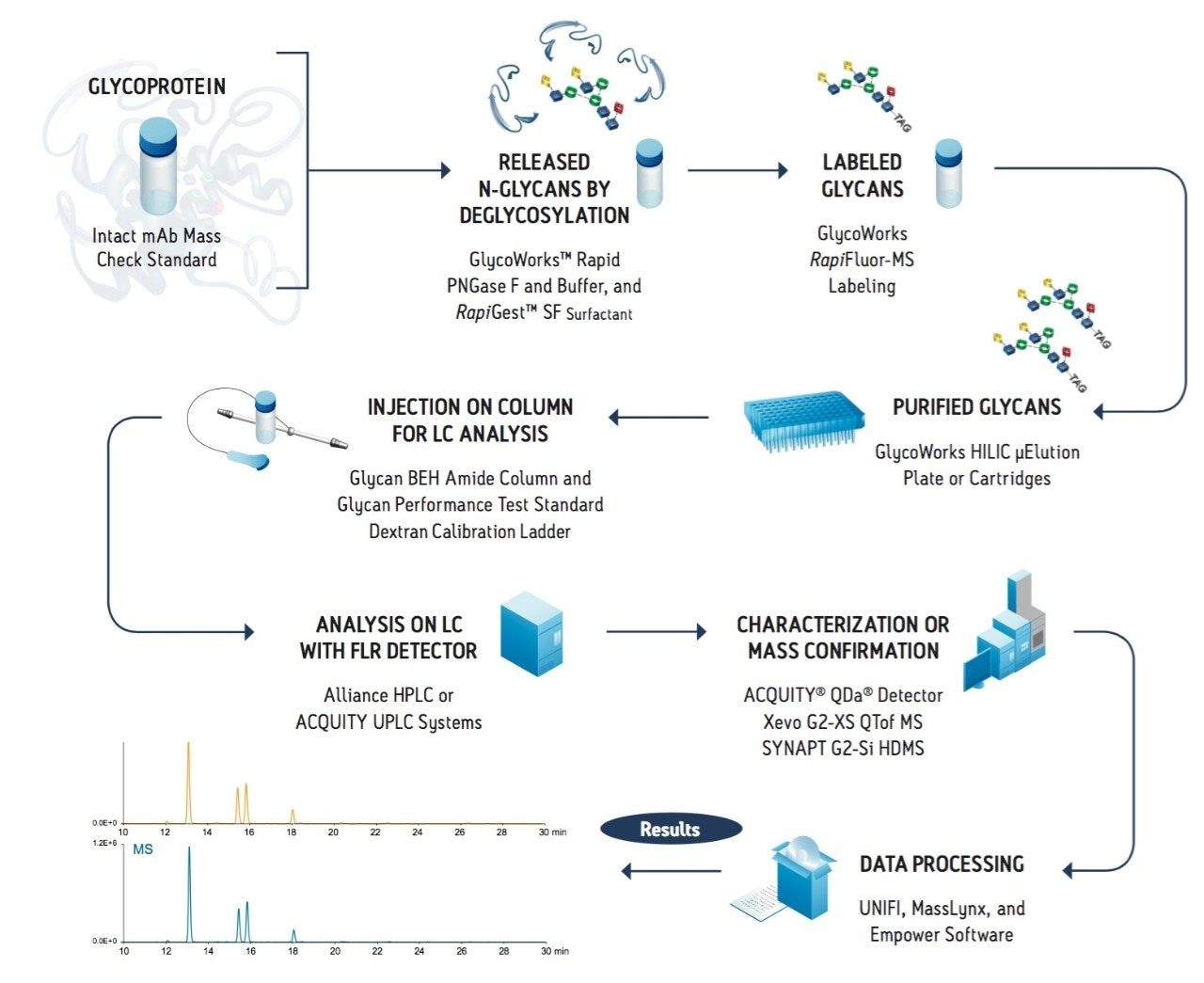
The GlycoWorks RapiFluor-MS N-Glycan Kit provides a novel formulation of Rapid PNGase F and RapiGest SF Surfactant that can be used to completely deglycosylate a diverse set of glycoproteins in an approximately 10 minute procedure. This fast deglycosylation procedure is facilitated by the use of RapiGest, an anionic surfactant, that is used to ensure that N-glycans are accessible to Rapid PNGase F and that glycoproteins remain soluble upon heat denaturation. Most importantly, RapiGest is an enzyme-friendly reagent and can therefore be used at high concentrations without hindering the activity of Rapid PNGase F. In the developed fast deglycosylation technique, a glycoprotein is subjected to a high concentration of RapiGest (1%) and heated to ≥80˚C for 2 minutes. Subsequently and without any additional sample handling, Rapid PNGase F is added to the solution and the mixture is incubated at an elevated, 50˚C temperature for 5 minutes to achieve complete, unbiased deglycosylation.
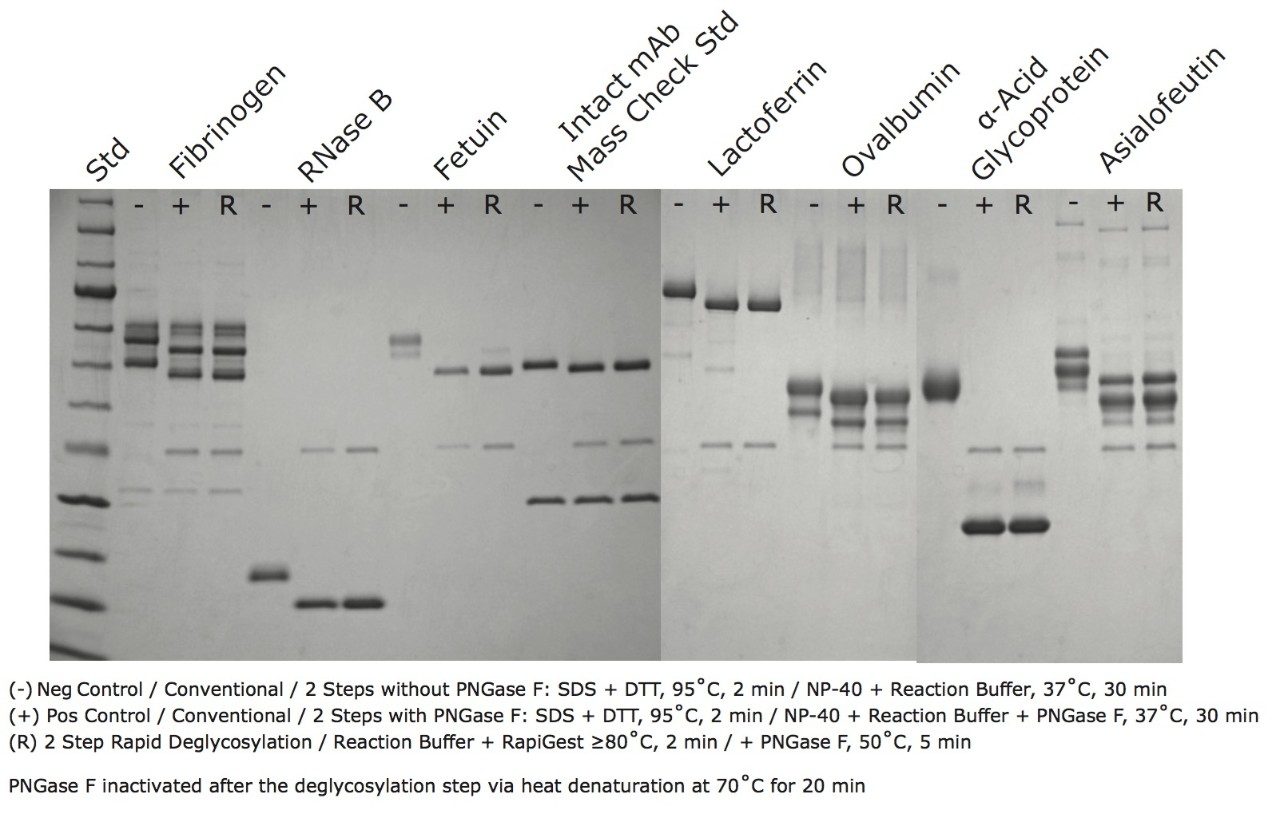
The effectiveness of this rapid deglycosylation process has been evaluated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE is an effective technique for separating proteins based on their size in solution and can often be used to separate the glycosylated and de-glycosylated forms of proteins.17-18 A diverse set of glycoproteins were deglycosylated according to the rapid deglycosylation procedure and analyzed by SDS-PAGE along with negative controls containing no PNGase F and positive controls, in which the glycoproteins were subjected to conventional multiple step deglycosylation with SDS based denaturation and PNGase F incubation for 30 minutes at 37 ˚C. Figure 7 shows the results of this study, where it can be seen that for each of the tested proteins there is a significant decrease in protein apparent molecular weight after they are subjected to the rapid deglycosylation procedure. Moreover, the apparent molecular weight decreases are visually comparable to those observed for proteins deglycosylated by the control method. These results demonstrate that the fast deglycosylation approach facilitated by a unique formulation of Rapid PNGase F and RapiGest SF Surfactant produces deglycosylation comparable to a conventional approach but in only a fraction of the time required.
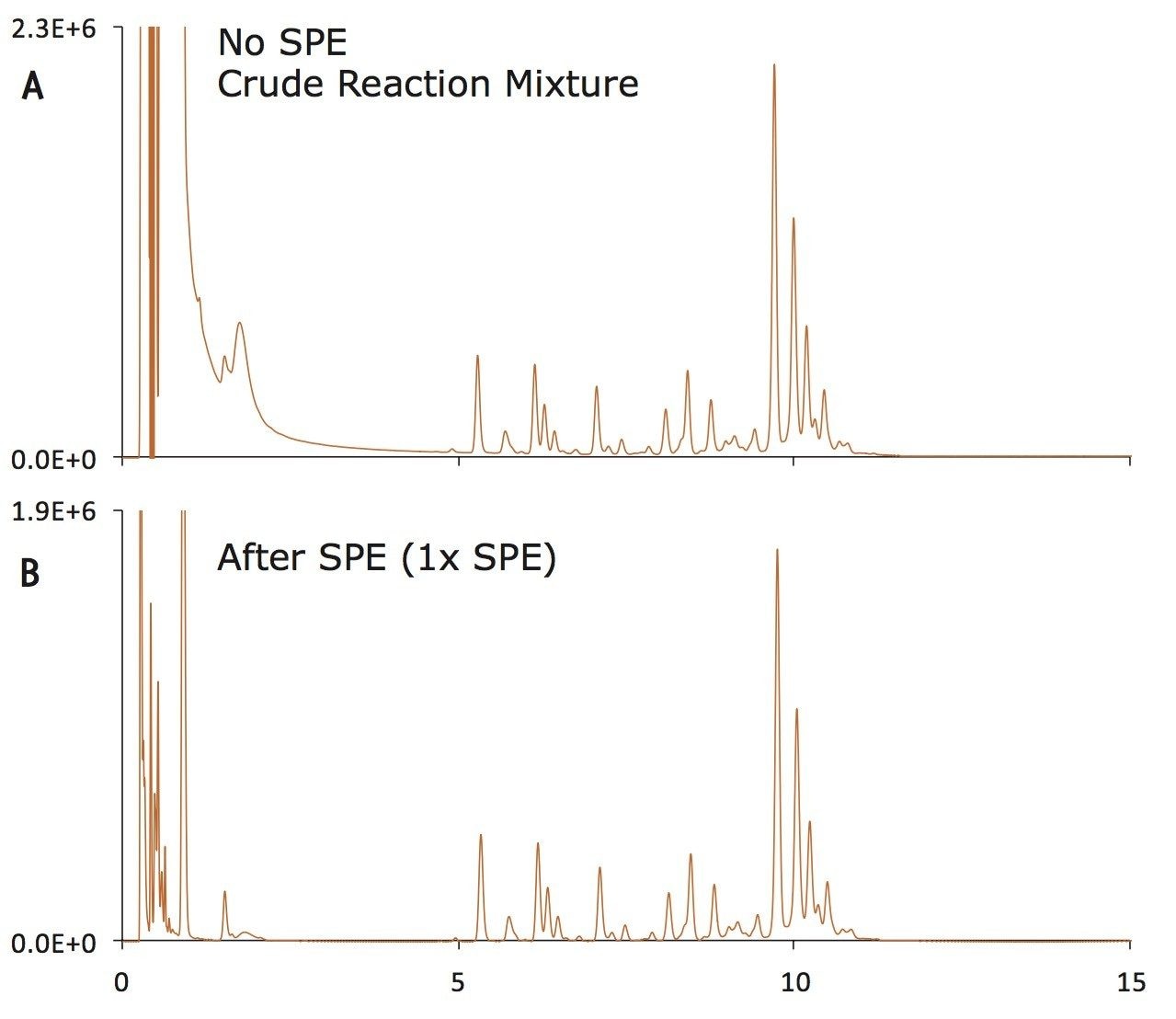
As mentioned earlier, the final step in an N-glycan sample preparation aims to extract the labeled glycans in preparation for their analysis. An effective approach for extraction of labeled glycans from reaction byproducts has been devised using solid phase extraction (SPE). In particular, this SPE is designed to selectively extract RapiFluor-MS labeled N-glycans from a mixture comprised of deglycosylated protein, PNGase F, buffer/formulation components, RapiGest Surfactant, and labeling reaction byproducts, which otherwise interfere with analysis of the labeled glycans by HILIC column chromatography (Figure 8). For the RapiFluor-MS N-Glycans kit, a GlycoWorks µElution plate is provided that contains a silica based aminopropyl sorbent specifically selected for this application.19-22 Due to its highly polar nature, the GlycoWorks SPE sorbent readily and selectively retains polar compounds such as glycans. In addition, this sorbent possesses a weakly basic surface that provides further selectivity advantages based on ion exchange and electrostatic repulsion. It is also worth noting that the GlycoWorks µElution Plate is designed for minimal elution volumes such that samples can be immediately analyzed without a dry down step. Moreover, the GlycoWorks µElution Plate is constructed as a 96 well format, meaning it can be used to perform high throughput experiments or used serially (with appropriate storage, see the Care and Use Manual) for low throughput needs.
In this HILIC SPE process, the sorbent is first conditioned with water and then equilibrated to high acetonitrile loading conditions. Thereafter, glycan samples that have been diluted with acetonitrile are loaded and washed free of the sample matrix using an acidic wash solvent comprised of 1% formic acid in 90% acetonitrile. This washing condition achieves optimal SPE selectivity by introducing electrostatic repulsion between the aminopropyl HILIC sorbent and reaction byproducts and by enhancing the solubility of the matrix components. After washing, the labeled, released glycans are next eluted from the HILIC sorbent. Since the GlycoWorks SPE sorbent has a weakly basic surface, and the capacity for anion exchange, just as it has the capacity for cation repulsion, it is necessary to elute the labeled glycans with an eluent of significant ionic strength. We have, as a result, developed an elution buffer comprised of a pH 7 solution of 200 mM ammonium acetate in 5% acetonitrile (p/n 186007991). Upon their elution, the RapiFluor-MS labeled glycans can be diluted with a mixture of organic solvents (acetonitrile and dimethylformamide) and directly analyzed by UPLC or HPLC HILIC column chromatography using fluorescence and/or ESI-MS detection.
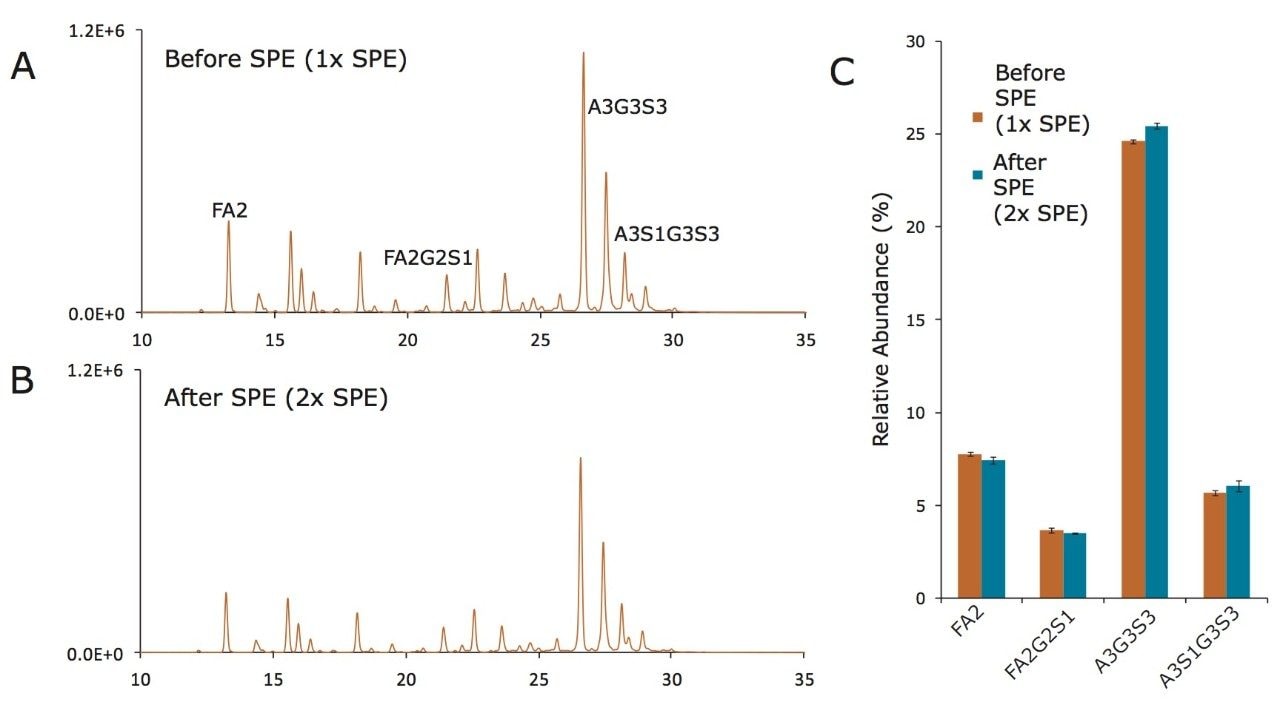
As with other aspects of the RapiFluor-MS N-Glycan Kit, this GlycoWorks SPE step has been extensively evaluated. A test mixture was created to assess the recovery of a diverse set of RapiFluor-MS labeled N-glycans, ranging from small neutral glycans to high molecular weight, tetrasialylated glycans. Such a mixture was prepared by releasing and labeling N-glycans from both pooled human IgG and bovine fetuin. An example analysis of this test mixture by HILIC column chromatography and fluorescence detection is shown in Figure 9A. Species representing extremes in glycan properties are labeled, including an asialo FA2 glycan and a glycan with a tetrasialylated, triantennary structure (A3S1G3S3). To evaluate the effects of the SPE process, this mixture was subjected to a second pass of GlycoWorks SPE and again analyzed by HILIC chromatography and fluorescence detection, as shown in Figure 9B. It can be observed that the sample processed twice by SPE presents a labeled glycan profile comparable to the profile observed for the sample processed only once by SPE. Indeed, this SPE step has been found to exhibit an absolute recovery of approximately 70-80% for all purified glycans and more importantly highly accurate relative yields. Figure 9C shows the relative abundances for four glycans (FA2, FA2G2S1, A3G3S3, and A3S1G3S3) as determined for samples subjected to one pass versus two passes of SPE. The largest deviation in relative abundance was observed for the tetrasialylated A3S1G3S3, in which case relative abundances of 5.7% and 6.1% were determined for samples processed by one and two passes of SPE, respectively. With these results, it is demonstrated that GlycoWorks SPE provides a mechanism to immediately analyze a sample of extracted, labeled glycans and does so without significant compromise to the accuracy of the relative abundances determined for a wide range of N-glycans
Conventional approaches to the preparation of N-glycans for HILIC-FLR-MS are either laborious, time-consuming, or require compromises in sensitivity. With the development of the GlycoWorks RapiFluorFluor-MS N-Glycan Kit, we address these shortcomings by enabling unprecedented sensitivity for glycan detection while also improving the throughput of N-glycan sample preparation. With this approach, glycoproteins are deglycosylated in approximately 10 minutes to produce N-glycosylamines. These glycans are then rapidly reacted with the novel RapiFluor-MS reagent within a 5 minute reaction and are thereby labeled with a tag comprised of an efficient fluorophore and a highly basic tertiary amine that yields enhanced sensitivity for both fluorescence and MS detection. In a final step requiring no more than 15 minutes, the resulting RapiFluor-MS labeled glycans are extracted from reaction byproducts by means of µElution HILIC SPE that has been rigorously developed to provide quantitative recovery of glycans (from neutral to tetrasialylated species) and to facilitate immediate analysis of samples. Accordingly, an analyst can complete an N-glycan sample preparation, from glycoprotein to ready-to-analyze sample, in just 30 minutes when using the sensitivity enhancing RapiFluor-MS labeling reagent.
720005275, January 2015