In this application note an LC-MRM assay is developed for flexible, high-throughput, sub-ppm quantification assays of multiple HCPs across multiple preparations of biopharmaceuticals.
Compared with the traditional process-specific ELISA assays used for quantification of individual host cell proteins (HCPs), the LC-MRM assays achieve similar sensitivities while they are much more flexible, faster and cheaper to develop.
Protein biopharmaceuticals are typically produced by recombinant DNA technology in well-selected cell lines. Even after extensive purification, low-level protein impurities (1–100 ppm) derived from the host cell line – HCPs, can still be present in the formulated drug substance (DS). The presence of HCPs in the biopharmaceutical product can have several detrimental effects, most concerning being the possibility to produce unwanted immunogenic effects in patients. In addition, HCPs can affect drug stability and efficacy or be responsible for drug degradation. For these reasons, regulatory agencies require that several HCP identification and quantification assays are performed prior to drug approval.
Phospholipase B-Like 2 (PLBL2) is a 65.5 kDa host-cell-protein (HCP) that co-purifies frequently in mAb preparations produced in Chinese hamster ovary (CHO) cells which was identified for the first time by mass spectrometry by a group of scientists from Roche/Genentech.1 The same protein or similar PLBL2 isoforms were later identified in mAb preparations by other researchers from Janssen,2 Eli Lilly and Company3 and Shire.4 The protein was shown to be responsible for enzymatic degradation of polysorbates 20 and 80, which are common adjuvants present in formulated DS.2-3 For this reason, PLBL2 affects DS (mAb) stability and needs to be removed from mAb preparations down to acceptable levels. The interactions between PLBL2 and mAbs were recently investigated to understand the factors responsible for the presence of PLBL2 following Protein A purifications.5
The acceptable levels tolerated for this HCP are in the sub ppm range.1-4 While the typical assays used for HCP quantification, like the general CHO ELISA kits or the process-specific immunoassays (e.g. ELISAs) are very sensitive (<1 ppm), they cannot provide individual HCP measurements. It is possible to develop ELISA assays for each individual HCP, but in reality this approach is not feasible to be implemented for each HCP impurity due to time constraints (>6 months) and development costs (>$500K). A process specific ELISA PLBL2 assay had to be developed in house recently1 because the commercial ELISA PLBL2 kit failed to detect the protein.
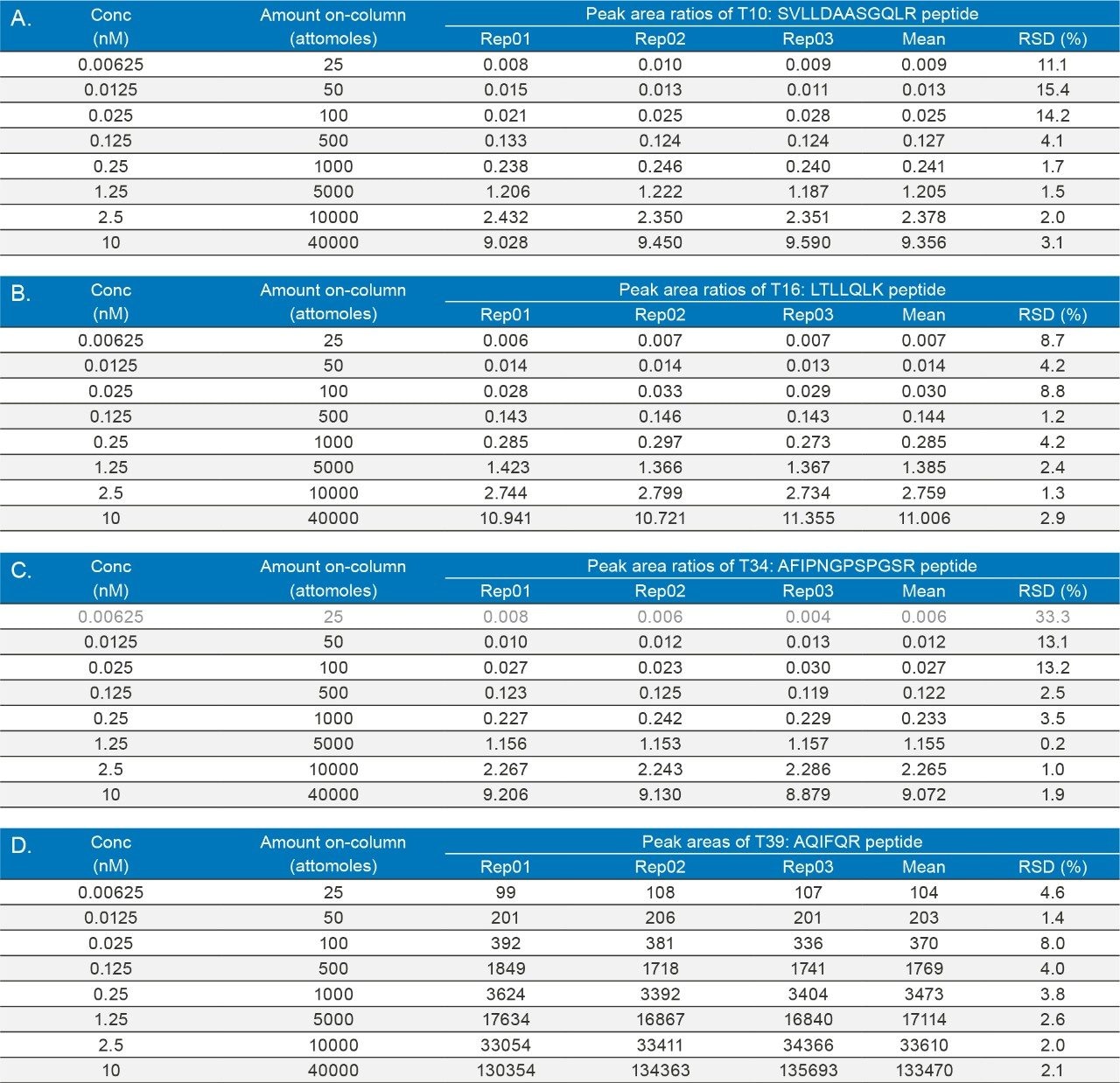
Another alternative to the process-specific PLBL2 ELISA assays, is the LC/MRM assay described here which was developed in significantly less time (one week) and with considerably less resources compared to the PLBL2 ELISA assay. The LC-MRM assay presented here is able to quantify PLBL2 from mAb preparations down to concentrations of 0.00625 nM (or 25 attomoles or 1.6 pg of PLBL2 digest loaded on-column). For a total mAb digest amount of 10 µg loaded on-column, the LOQ of the PLBL2 assay developed here is ~160 ppb (part-per-billion). The ppb level quantification of PLBL2 was confirmed with four tryptic peptides using their corresponding MRM transitions. Peak area relative standard deviation (RSD) for replicate measurements (n=3) was better than 15% when using the corresponding 13C15N-isotopically labeled peptide internals standard. In addition, the assay had a wide dynamic range of at least three orders of magnitude – 1600. Assay robustness was demonstrated by performing 90 injections for a concentration near the LOQ level (0.025 nM PLBL2 digest or 100 attomoles protein digest loaded on-column) when the peak area RSDs were also better than 15% for all four peptides.
Clearly the LC-MRM assay described here opens the door for flexible, high-throughput, sub-ppm quantification assays of multiple HCPs across multiple preparations of biopharmaceuticals.
A highly purified mAb (NIST mAb candidate reference LRM 8670) produced in a murine suspension cell culture was acquired from the National Institute of Standards and Technology (NIST) at a concentration of 10 mg/mL. The NIST mAb was denatured with 0.05% RapiGest surfactant (60 °C, 15 min), reduced with 25 mM DTT (60 °C, 1h), alkylated with 10 mM IAM (RT, 30 min in the dark) and digested with porcine trypsin (Promega, Madison, WI, USA)) overnight (16 h, 37 °C) using a 10:1 molar ratio of mAb : enzyme. After digestion, the RapiGest surfactant was decomposed by adding 5 µL of formic acid (FA, Sigma-Aldrich, St. Louis, MS, USA) and the digest was incubated for 30 min at 37 °C and centrifugated (15 min, 4,000 rpm) to separate the insoluble component of RapiGest by precipitation. The supernatant was then transferred to a TruView LCMS certified clear glass vial (P/N 186005663CV). LC/MS-grade organic solvents (acetonitrile-ACN, isopropanol-IPA and methanol-MeOH) were purchased from Thermo Fisher Scientific.
Since the hamster (CHO) PLBL2 protein was not commercially available, the rat PLBL2 protein, obtained from MyBioSource Inc. (San Diego, CA, USA), was used instead. The rat PLBL2 protein (0.2 mg with a concentration of 5 mg/mL) was digested with trypsin following the NIST mAb protocol described above.
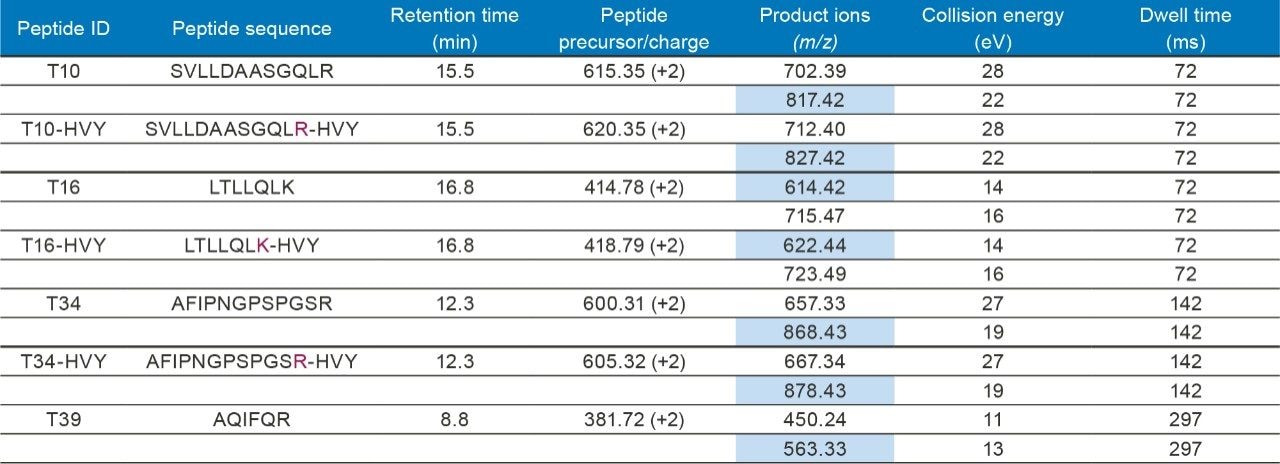
All seven tryptic peptides used for PLBL2 quantification were obtained from New England Peptide (Gardner, MA, USA): four light (non-isotopically labeled) peptides (T10, T16, T34, and T39, see Figure 1 and Table I) and three heavy (13C15N-isotopically labeled) peptides (T10-HVY, T16-HVY, and T34-HVY, see Figure 1 and Table I as well). The light peptides were spiked in the NIST mAb digest at eight different concentrations: 0.00625; 0.0125; 0.025; 0.125; 0.25; 1.25; 2.5, and 10 nM (or fmoles/µL), while the heavy peptides were used as internal standards at a fixed concentration of 1 nM (or 1 fmole/µL). With an injection volume of 4 µL, the amounts of light peptides loaded on-column for the 8 concentrations mentioned above were: 25; 50; 100; 500; 1000; 5000; 10000 and 40000 attomoles, while the amount of NIST mAb digest was kept constant at 10 µg for every injection. The amount of each heavy peptide was also kept constant for every injection (4 fmoles on-column). Sample blanks, containing only the NIST mAb digest without the spiked PLBL2 peptides, were prepared as well.

|
LC system: |
ACQUITY UPLC I-Class |
|
Column: |
1.0 x 150 mm, packed with 1.7 μm CSH C18 particles (P/N 186005294) |
|
Column temp.: |
60 °C |
|
Flow rate: |
50 μL/min |
|
Mobile phases: |
Solvent A: 0.1% FA in DI water |
|
Solvent B: 0.1% FA in acetonitrile |
|
|
Injection volume: |
4 μL, partial loop injection using a 10 μL injection loop |
|
Wash solvents: |
SNW (1:1:1:1 IPA:ACN:MeOH:H2O with 0.2% FA) WNW (0.1% FA in H2O) |
|
Time (min) |
Flow rate (µL/min) |
Solvent A composition (%) |
Solvent B composition (%) |
Gradient curve |
|---|---|---|---|---|
|
0 |
50 |
98 |
2 |
6 |
|
2 |
50 |
98 |
2 |
6 |
|
20 |
50 |
75 |
25 |
6 |
|
21 |
50 |
10 |
90 |
11 |
|
26 |
50 |
98 |
2 |
11 |
|
35 |
50 |
98 |
2 |
11 |
|
MS system: |
Xevo TQ-S Tandem Quadrupole |
|
Mass Spectrometer Ionization mode: |
ESI (+) |
|
ESI probe: |
Low flow probe (P/N 186007529) |
|
Capillary voltage: |
3.0 kV |
|
Cone voltage: |
35 V |
|
Desolvation temp.: |
600 °C |
|
Desolvation flow: |
1000 L/hr |
|
Nebulizer gas flow: |
7 bar |
|
Cone gas flow: |
150 L/hr |
|
Collision cell pressure: |
3.83 e-3 Torr |
|
Data acquisition software: |
MassLynx 4.1 SCN 950 |
|
Quantification software: |
TargetLynx |
Hamster phospholipase B-Like 2 (PLBL2) was recently identified as a low-level HCP contaminant in several high-purity mAb preparations produced in several CHO cell lines.1-4 The full protein sequence, containing 585 AA, is displayed in Figure 1. Four tryptic peptides (highlighted in red) were selected for LC-MRM quantification based on high-resolution peptide mapping data obtained for the rat PLBL2 digest. Four stable proteotypic peptides, highlighted in Figure 1, were selected based on the strong response of their precursors in ESI-MS. The LC-MRM signals were individually optimized for each peptide transition in terms of collision energy and the MRM/SRM experimental conditions of the assay are summarized in Table I.
As shown in Figure 2, all four tryptic PLBL2 peptides monitored (T10, T16, T34, and T39) were detected with similar sensitivities at the lowest spiked concentration: the LOQ concentration was 0.00625 nM, corresponding to 25 attomoles of PLBL2 peptides loaded on-column. Also shown in the same figure are the LC-MRM chromatograms recorded for another concentration, near the LOQ level: 0.0125 nM or 100 attomoles PLBL2 peptides loaded on-column.

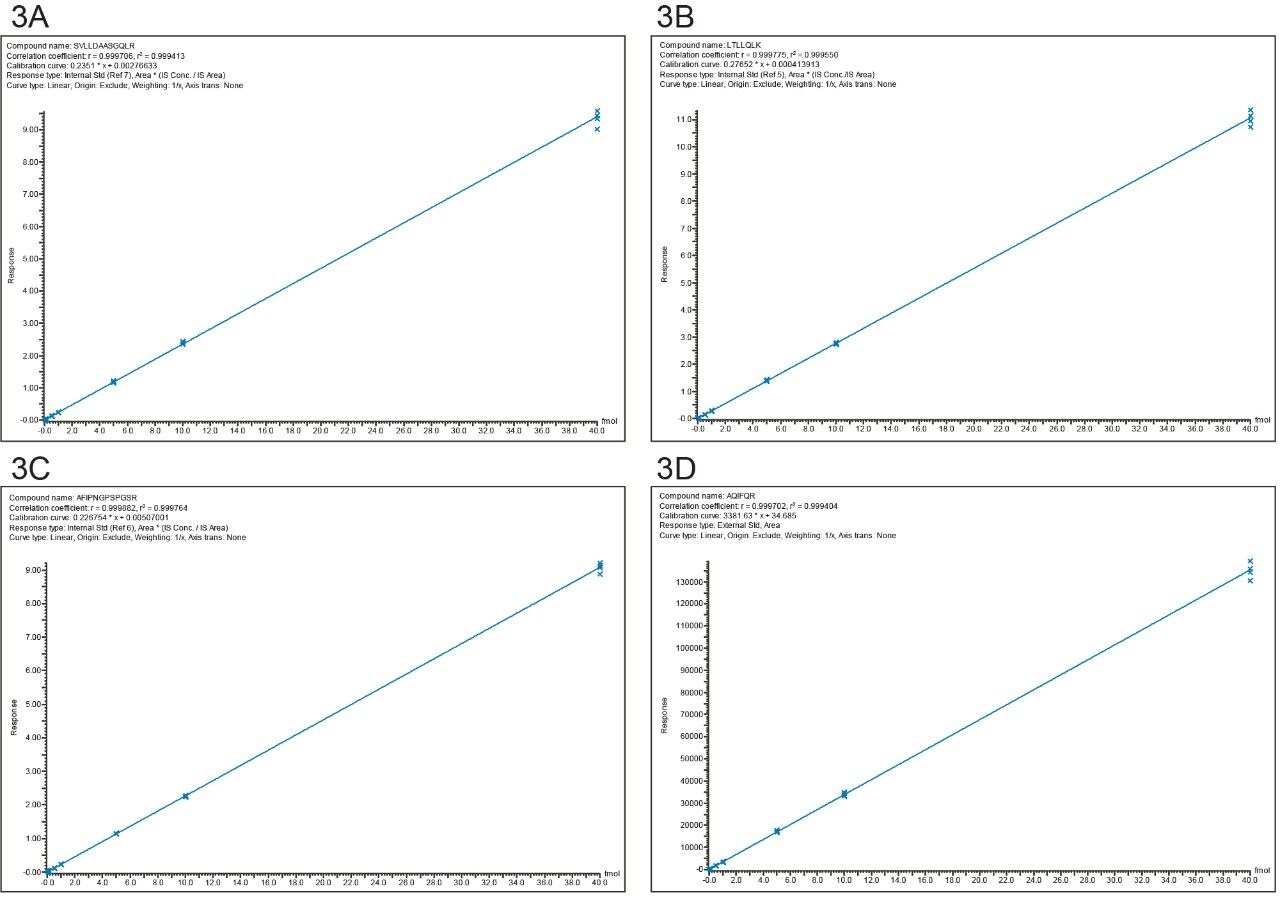
The reproducibility of the assay is illustrated by the data presented in Table II which shows the results obtained for replicate injections (n=3) across all eight concentrations investigated. Individual TargetLynx calibration curves obtained for each peptide are presented in panels A–D of Figure 3.
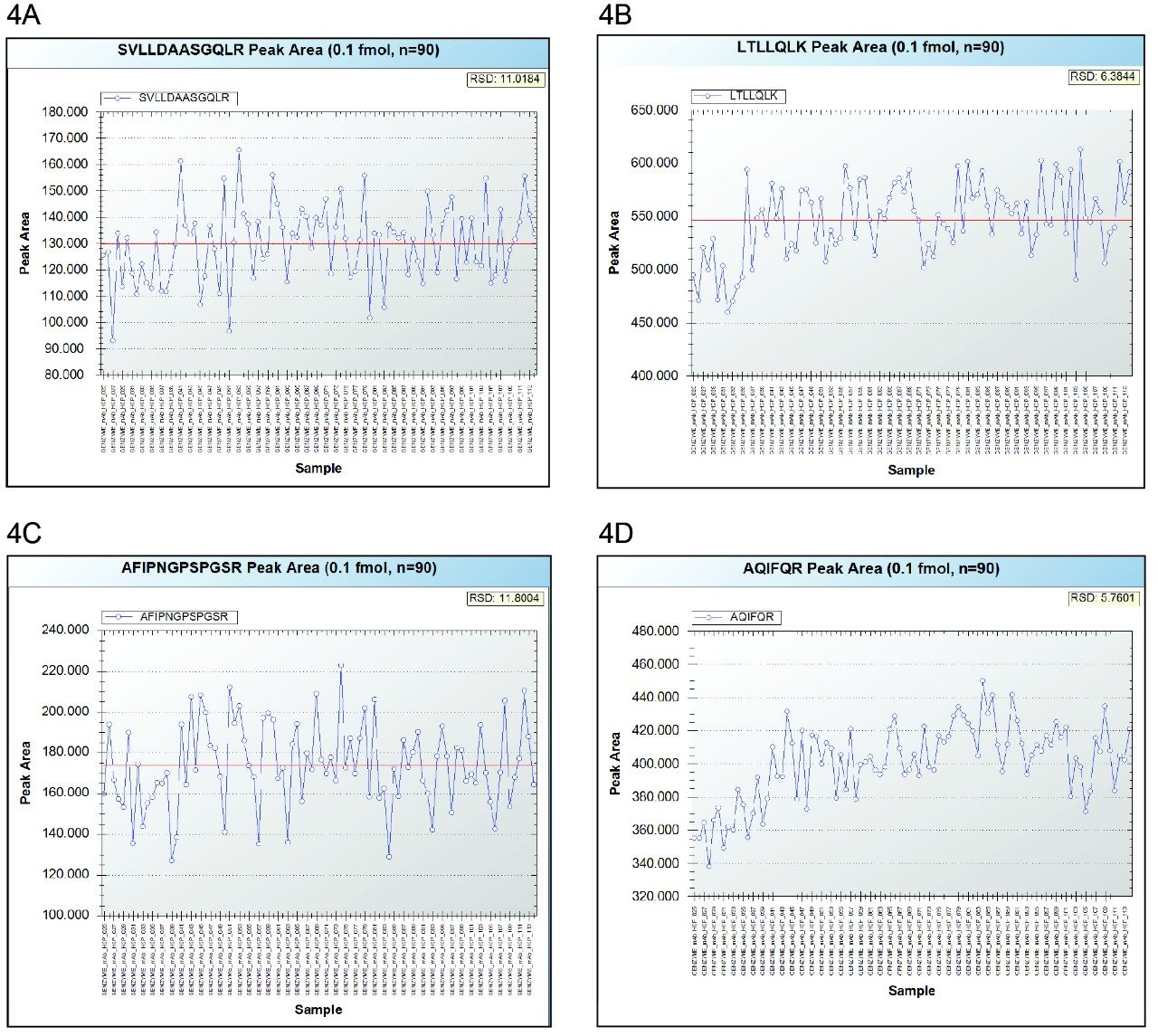
In addition, the robustness of the assay was tested with 90 injections performed near the LOQ level (100 attomoles on-column for each peptide) and the peak area trend plots are shown in Figure 4.
The exquisite sensitivity of the PLBL2 LC-MRM assay is highlighted by the chromatograms shown in Figure 2. All four peptides (T10, T16, T34, and T39), carefully selected to cover several critical protein domains, were detected at the lowest spiked concentration (0.00625 nM, 25 attomoles loaded on-column) with S/Ns in the range of 5–10. In addition, as indicated by the blank chromatograms recorded for the free (not spiked) NIST mAb digest, the assay was free of significant MRM/SRM interferences even in the presence of a significant amount (10 µg) of peptide matrix. The LOQ of the assay for three peptides (T10, T16, and T39) was the same as the LOD, indicating the ability of the LC-MRM assay to measure PLBL2 in mAb preparations down to ~160 ppb (part-per-billion). For the fourth peptide (T34, Figure 2C), the LOQ of the assay was 0.0125 nM (50 attomoles on-column), which corresponds to ~360 ppb. The reproducibility of the assay (shown in Table II), which was tested by replicate injections (n=3) across the entire concentration range investigated (0.00625–10 nM, 1600 dynamic range), was better than 15% at all concentrations at and above the LOQ.
It is worth mentioning that the original work that led to the mass spectrometric identification of CHO PLBL2 in mAb preparations was triggered by nonlinear dilution effects observed in the original total CHO protein (CHOP) ELISA assay.1 This general CHOP assay was able to measure PLBL2 accurately at low concentrations (10 ng/mL or ~10 ppm), but at higher concentrations (300 ng PLBL2 per mg mAb or 300 ppm) it failed to provide reliable measurements. By contrast, the LC-MRM assay presented here has the appropriate dynamic range for PLBL2 measurements. At least three orders of magnitude are demonstrated by the four calibration curves displayed in Figure 3.
Finally, the assay robustness was evaluated by 90 consecutive injections performed at a concentration level near the LOQ (0.025 nM PLBL2 digest or 100 attomoles protein digest loaded on-column). The trend plots shown in Figure 4 indicate that the peak area RSDs were also better than 15% for all four peptides.
The authors would like to thank Matthew Stone (Waters Corporation) for providing several useful insights in the initial phase of this project.
720006162, December 2017