A Simple, Broadly Applicable Automated Bioanalytical Sample Preparation Strategy for LC-MS Quantification of Apixaban: Evaluation of Common Bioanalytical Extraction Techniques
Abstract
The following work demonstrates a simple, generic, broadly applicable, fully automated sample preparation strategy, requiring no method development, for several common bioanalytical extraction techniques including protein precipitation (PPT), supported-liquid extraction (SLE), and solid phase extraction (SPE). All sample preparation and extraction was performed on the Andrew+ Pipetting Robot using generic extraction protocols, for the subsequent LC-MS/MS detection, and quantification of therapeutic drug apixaban from plasma.
Benefits
- Simplified bioanalytical sample preparation strategy, requiring no method development for successful analyte extraction from biomatrices using PPT, LLE, SLE, and SPE
- Generic extraction method protocols, yielding high analyte recovery and achieving reproducible results, from extracted plasma
- Automated sample extraction, for “walk-away” method execution, mitigating risk of manual error, improving analytical performance and freeing up scientist time
- Excellent quantitative performance, with linear dynamic range from 2–500 ng/mL, QC accuracies ≥90% with RSDs ≤10% for apixaban extracted from plasma
Introduction
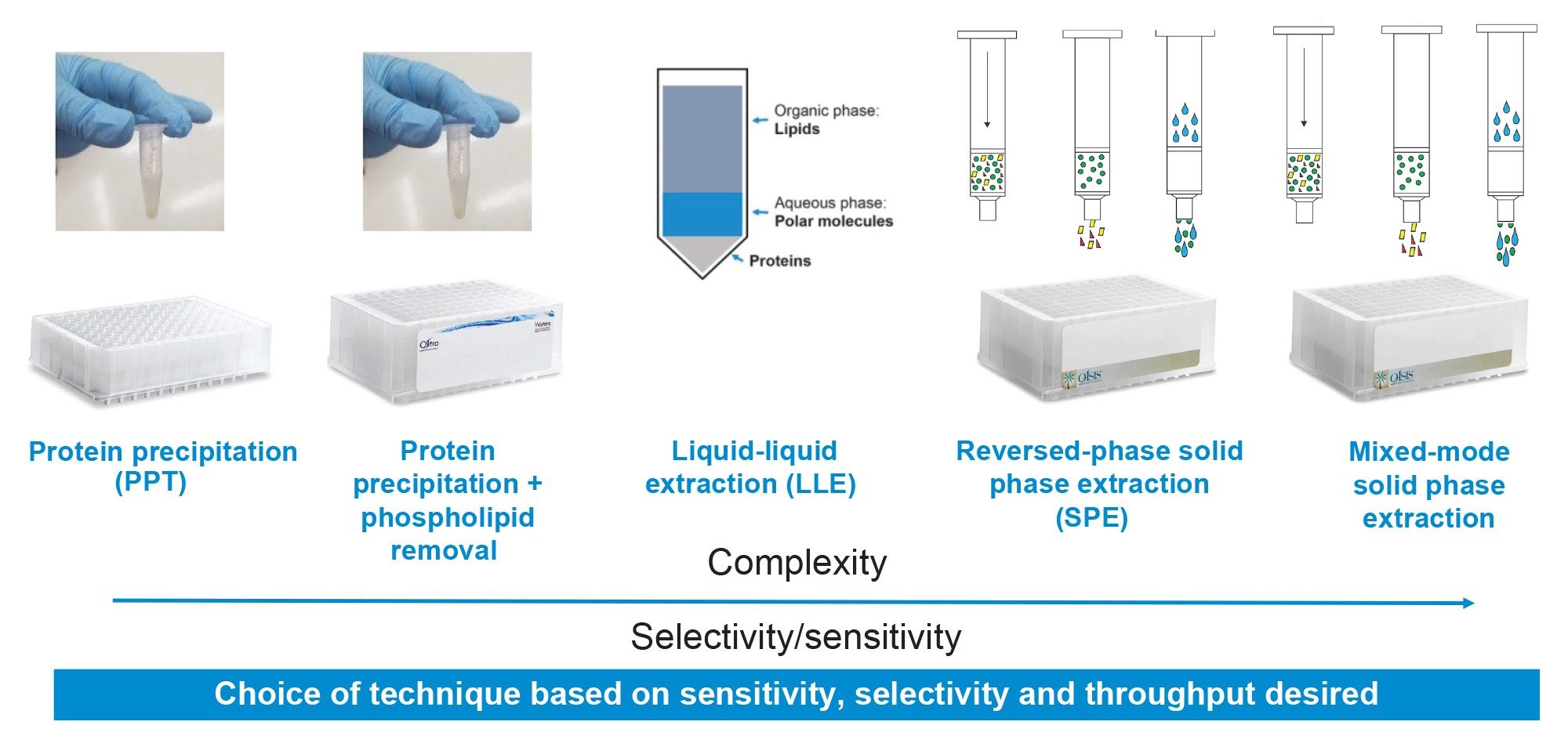
Bioanalytical methods are essential to support discovery and development of new pharmaceutical candidates. Common sample preparation preparation methods can range from simple techniques such as dilute and shoot or protein precipitation (PPT) to more targeted and specialized methods such as liquid-liquid extraction (LLE), solid phase extraction (SPE) or immunoaffinity purification. Regardless of the extraction technique, sample preparation and extraction of the pharmaceutical from biomatrices is often a bottle neck in the bioanalytical workflow, carrying the most complexity and method development time due to the variety of extraction techniques that must be assessed, and the complex and laborious multi-step protocols associated with these techniques. Generally, the simpler techniques have wider applicability and require minimal method development with a trade-off of limited cleanliness and sensitivity. The more specific techniques offer superior cleanliness, specificity and sensitivity but have more limited applicability and may require more method optimization. The choice of sample preparation methods often depends on the application, biomatrices, and analytical requirements of the method. For general screening or qualitative work, a simple method such as sample dilution or protein precipitation (PPT) may be appropriate, while quantitative confirmation requiring high sensitivity often requires more specialized methods. This trend is highlighted in Figure 1, showing increasing complexity and specificity of different sample preparation techniques.
Regardless of the sample preparation technique used, it needs to be fit-for-purpose, achieving the required sensitivity, robustness and throughput. For bioanalytical liquid chromatography and mass spectrometry (LC-MS) methods, two key attributes that need to be evaluated are analyte recovery and matrix effects. Recovery is simply extraction efficiency, while matrix effects quantify the degree of ion suppression for a particular analyte within a specific method and is often used as a measure of extraction cleanliness. Additionally, monitoring the presence or degree of residual phospholipids can be used as a measure of sample cleanliness. Phospholipids (PLs) are one of the most common contributors to ion suppression in biological samples and minimizing them can improve the cleanliness and reliability of sample preparation techniques.
In this work, several common bioanalytical techniques including PPT, PPT with PL removal, supported liquid extraction (SLE), reversed-phase (RP) SPE, RP-SPE with PL removal and mixed-mode SPE were evaluated for the extraction of the therapeutic drug, apixaban from plasma. All extraction techniques were evaluated using generic manufacturer recommended protocols. All sample preparation and extraction was executed using the Andrew+ Pipetting Robot configured with the Extraction+ Connected Device, and controlled using its intuitive cloud-based OneLab Software. The automated sample protocols were adapted from existing OneLab Library methods, enabling easy execution. Analyte recovery, matrix effects and residual PLs were used to compare efficiency and cleanliness of the methods. Apixaban quantitative performance (e.g., linearity, accuracy, and precision) was also evaluated for each extraction technique. The results showed a general trend of increasing recoveries and decreasing matrix effects with increasing sample preparation specificity as we have seen in previous work (720005495 , 720006516).1,2
Experimental
Materials
Apixaban was purchased from Cerilliant (www.cerilliant.com). 13C-d3 Apixaban, used as internal standard (IS) was obtained from Cayman Chemicals (www.caymanchem.com). Stock solutions (1 mg/mL) were prepared in methanol (MeOH). Working stock solutions (10 µg/mL) were also prepared in methanol and were used to prepare calibrators and QC samples in plasma. Rat plasma (K3EDTA) was purchased from Innovative Research (www.innov-research.com). Daily working solutions for curve and QC generation were prepared in plasma. Calibration curves ranged from 2–500 ng/mL and QC samples were prepared at 4, 40, and 400 ng/mL. Plasma calibration standards and QC samples were prepared in triplicate (N=3). All recovery and matrix effects experiments were performed with 100 ng/mL concentrations of apixaban. LC-MS grade formic acid (FA) and phosphoric acid were purchased from Sigma Aldrich (www.sigmaaldrich.com). Tert-butyl methyl ether (MTBE) was obtained from Avantor sciences (www.avantorsciences.com). Methanol and Acetonitrile were purchased from Honeywell (lab.honeywell.com).
Sirocco Protein Precipitation plates, Ostro Protein Precipitation and Phospholipid Removal Plates, Oasis HLB, Oasis PRiME HLB, Oasis Method Development Sorbent Selection Plates, and Oasis MCX 96-well plates were all obtained from Waters. The Supported Liquid Extraction (SLE) plates (p/n: 96260-1) were obtained from Analytical Sales and Services (analytical-sales.com).
Recovery and Matrix Effects Calculations
Analyte recovery was calculated according to the following equation:
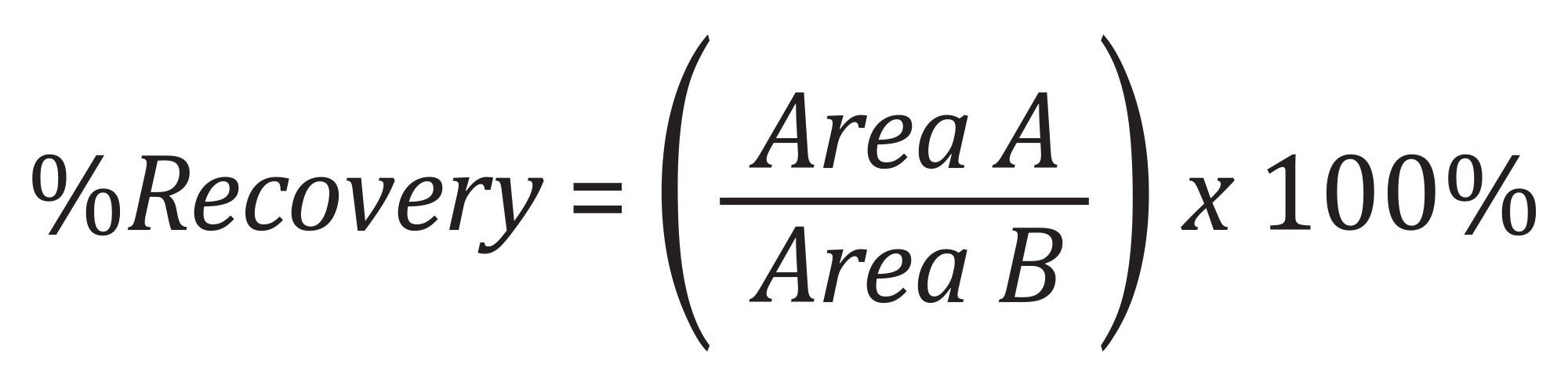
Matrix effects were calculated according to the following equation:
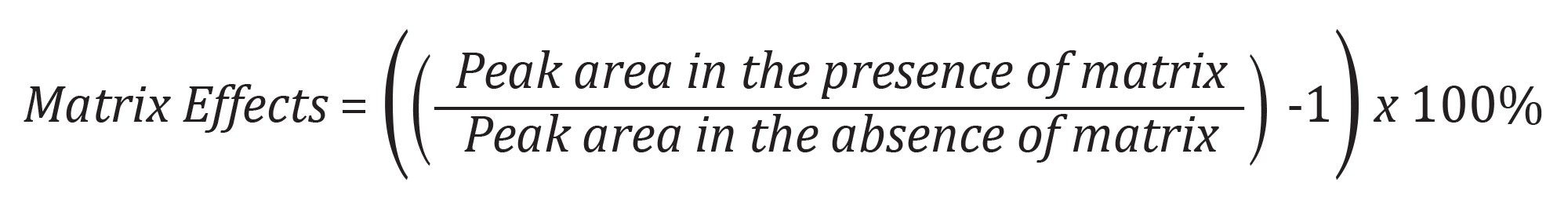
Automation Platform
The Andrew+ Pipetting Robot, equipped with the new Extraction+ Connected Device and controlled with the cloud-based OneLab Software, was used to design and execute the sample preparation and bioanalytical extraction protocols.
Extraction protocols
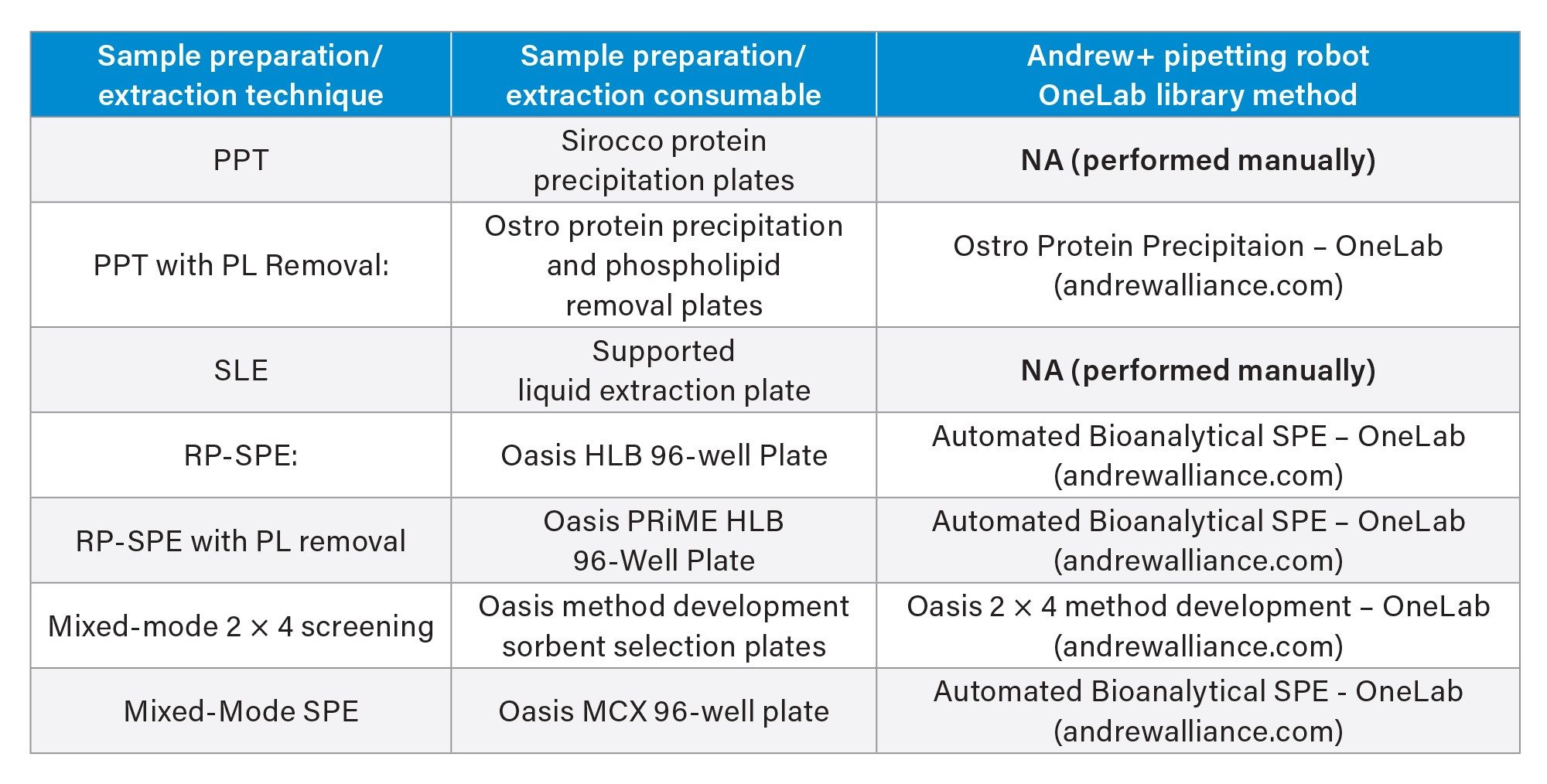
The OneLab Library protocols used for each technique are listed in Table 1 and graphical diagrams of the protocols used for each sample preparation method are shown in Figure 2. In each case, the manufacturers’ instructions for appropriate volumes and solvents were followed. All steps were fully automated by the Andrew+ Pipetting Robot, with the exception of the vortexing step for the PPT protocol and the evaporation step for the SLE protocol. Standard OneLab protocols from the OneLab Library https://onelab.andrewalliance.com/app/lab/D8xeYomN/library were downloaded and used for Ostro, Oasis HLB, Oasis HLB PRiME, and the Mixed-mode screening protocol. The 2 x 4 method development protocol also included steps for spiking the extracted samples to assess analyte recovery. New protocols were created for the PPT preparation using the Sirocco plate and the SLE plate.
OneLab Protocols

LC Conditions
|
LC system: |
ACQUITY I-Class UPLC (FL) |
|
Mobile phase-A: |
0.1% Formic Acid in 100% MilliQ water |
|
Mobile phase-B: |
0.1% formic Acid in 100% Acetonitrile |
|
Weak wash solvent: |
Water:Methanol (90:10 v/v) |
|
Strong wash solvent: |
Acetonitrile: Isopropanol: Water: Methanol (25:25:25:25 v/v/v/v) |
|
Detection: |
Xevo TQ-XS Mass Spectrometer |
|
Column(s): |
ACQUITY UPLC BEH C18 Column, 1.7 µm, 2.1 mm x 50 mm (p/n: 186002350) |
|
Column temperature: |
35 °C |
|
Column temperature: |
10 °C |
|
Injection volume: |
5 µL |
|
Flow rate: |
0.5 mL/min |
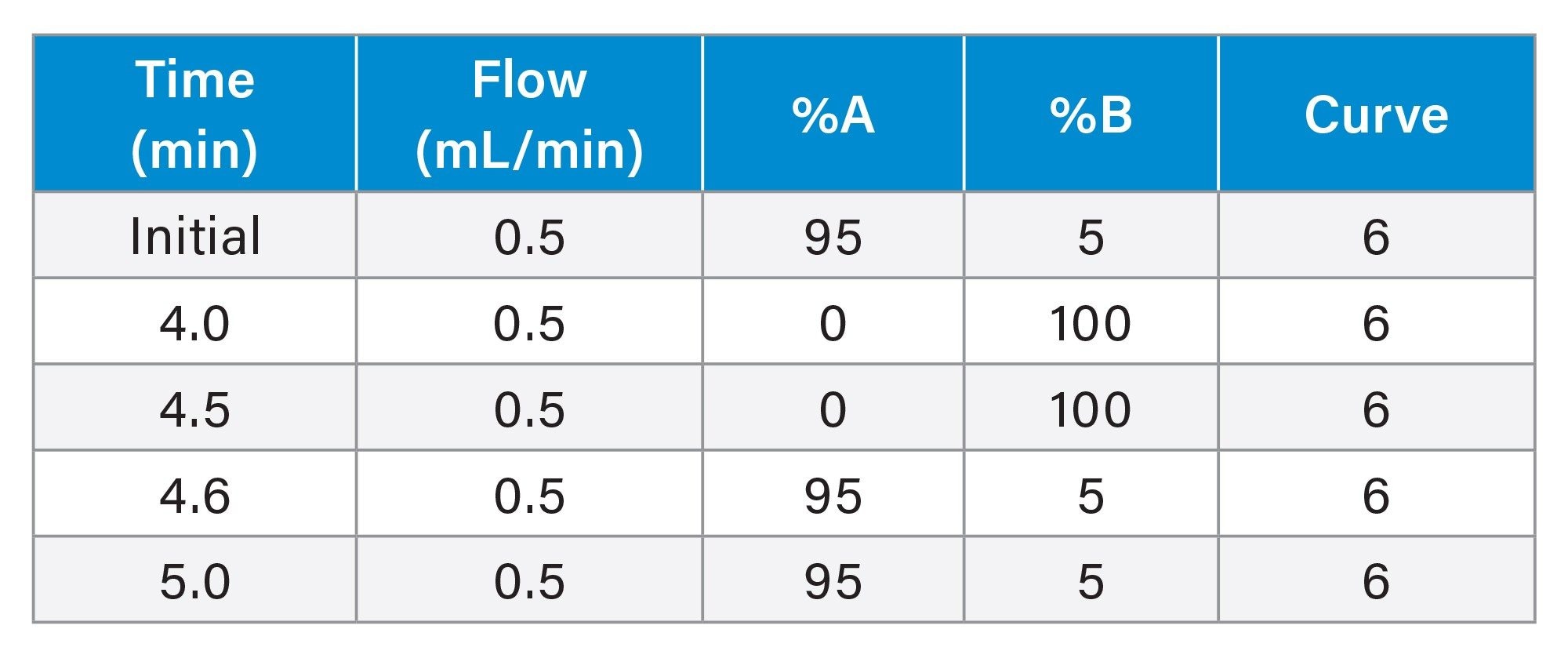
LC Gradient
MS Conditions
|
MS system: |
Xevo™ TQ-XS |
|
Ionization mode: |
ESI+ |
|
Acquisition range: |
MRM |
|
Capillary voltage: |
2.0 kV |
|
Cone voltage: |
30 V |
|
Desolvation temp: |
500 °C |
|
Desolvation flow: |
1100 L/Hr |
|
Cone gas flow: |
150 L/Hr |
|
Collision gas flow: |
0.2 mL/min |
|
Nebulizer gas flow: |
7 Bar |
Data Management
|
Instrument control software: |
MassLynx™ (v4.2) |
|
Quantification software: |
TargetLynx™ |
LC-MS Analysis
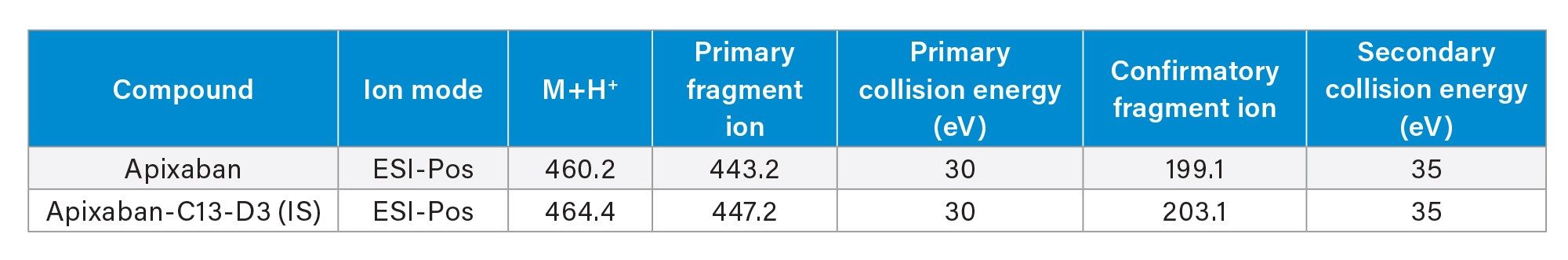
The chromatographic separation of apixaban was performed using a Waters ACQUITY I-Class UPLC and ACQUITY UPLC BEH C18 Column (1.7 µm, 2.1 x 50 mm) with gradient elution using water and acetonitrile mobile phases containing 0.1% formic acid. The flow rate was set at 0.5 mL/min with the column temperature set at 35 °C. Detection of analytes was performed with a Waters Xevo TQ-XS Mass Spectrometer in ESI+ mode using Multi Reaction Monitoring (MRM). The MS conditions for apixaban and its IS, apixaban C13-C3 are listed in Table 2.
Phospholipid Monitoring
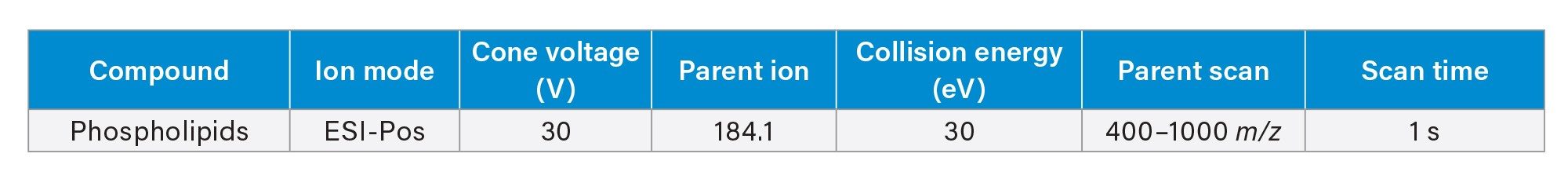
Residual phospholipids were analysed using the same UPLC gradient as the apixaban analysis. The MS conditions are shown in Table 3.
Results and Discussion
Automation
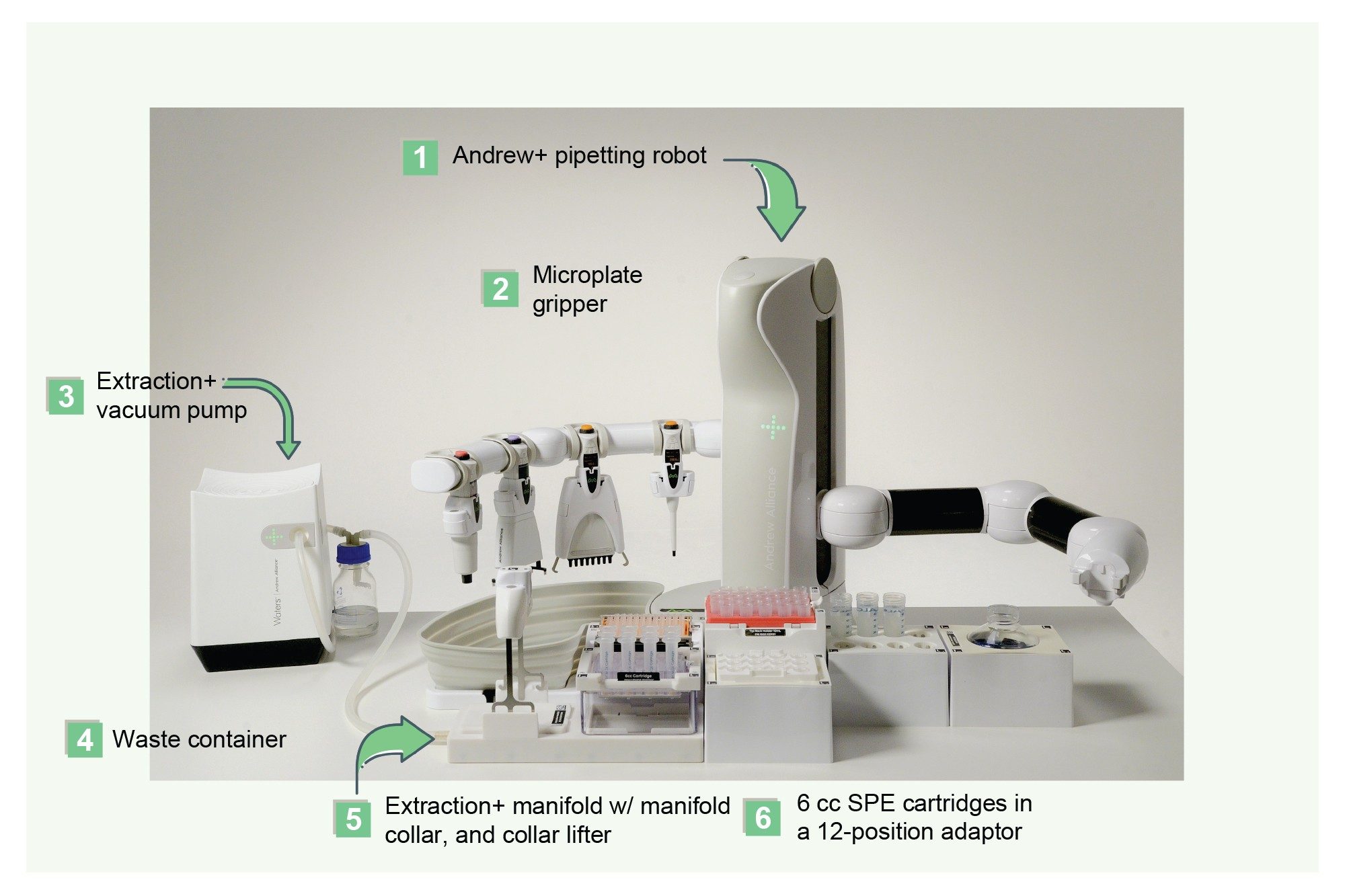
The Andrew+ Pipetting Robot configured with the Extraction+ Connected Device was used to extract the pharmaceutical, Apixaban, from plasma samples using a variety of common sample preparation techniques described above and in the protocols illustrated in Figure 2. All pipetting, reagent additions, sample pre-treatment and extraction device manipulations were fully automated, with manual intervention for the capping and vortexing step for the Sirocco plates and the solvent evaporation step during the SLE extraction. The Extraction+ Connected Device with flow-through waste collection and automatic placement of the sample extraction plates and collection labware onto the Extraction+ manifold enabled fully automated sample extraction. Figure 3 shows the Andrew+ Pipetting Robot configured with the Extraction+ Connected Device. The Andrew+ Pipetting Robot configured with Extraction+ Connected Device was used to perform initial method evaluation experiments, such as determining recovery and matrix effects, as well as quantitative extractions of calibration curves and QC samples.
Andrew+ Pipetting Robot OneLab Extraction Protocols
Figure 4 shows an example of the user interface generated by the OneLab Software. This particular OneLab protocol is for the SPE extraction of apixaban using the Oasis HLB 96-well plates and was created using the Automated Bioanalytical SPE Library method listed in Table 1. It shows the OneLab created deck layout for the Andrew+ Pipetting Robot and its protocol showing the position of all Dominos and connected devices. Similar lists and layouts were created for the other OneLab extraction protocols.

Chromatography
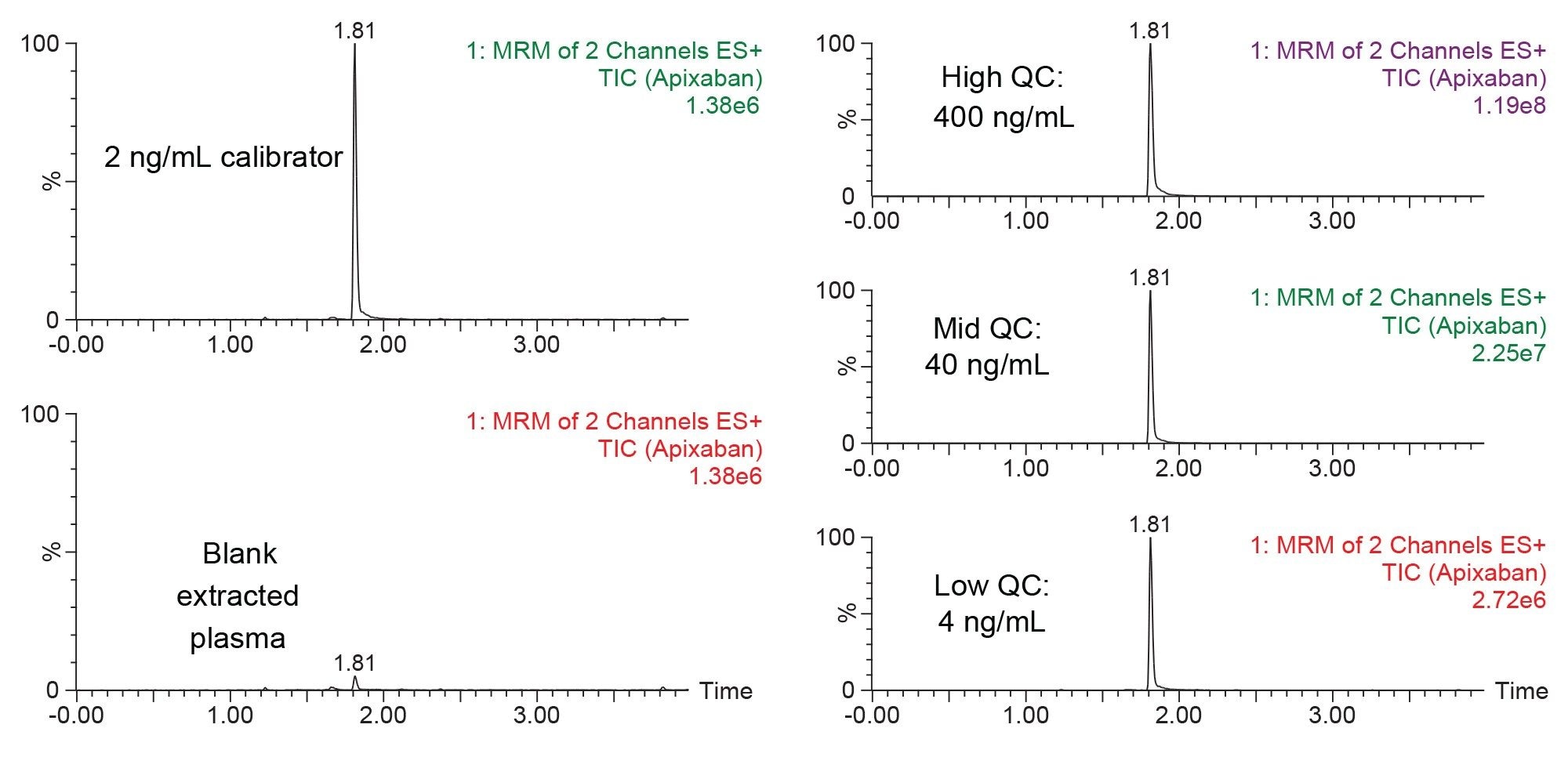
Figure 5 shows the chromatographic separation of apixaban under the LC conditions described in the experimental section. Panel A shows the low calibrator std (2 ng/mL) extracted plasma sample as compared to a blank extracted plasma sample, while Panel B highlights the chromatographic performance of the 3 quality control (QC) extracted plasma samples at concentrations of 4, 40, and 400 ng/mL apixaban. The ACQUITY UPLC BEH C18 Column (1.7 µm, 2.1 mm x 50 mm) provided consistent chromatography with no endogenous interferences. Carryover was minimal, with method blanks <10% the peak intensity of the lowest calibrators.
Recovery and Matrix Effects
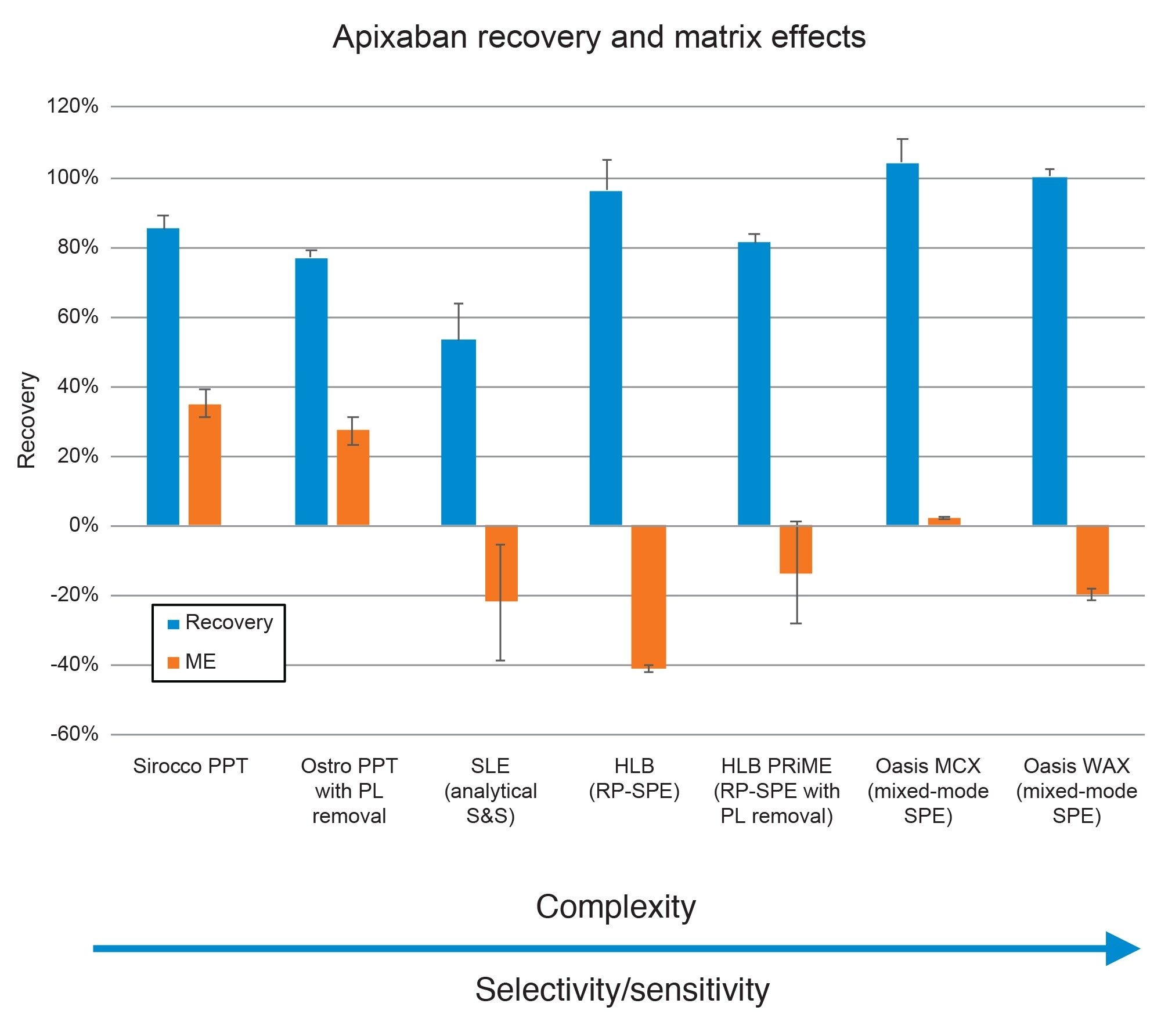
A key step in any bioanalytical procedure is the evaluation of extraction efficiency and cleanliness. This is done by calculating recovery and matrix effects for the target analytes as described in the Materials and Methods Section. Figure 6 shows the recovery and matrix effects (ME) results from the various sample preparation procedures. The sample preparation techniques are ordered by increasing selectivity, starting with the more universal methods such as protein precipitation and progressing to the more selective and specific mixed-mode SPE procedures. A general trend of improved recovery and decreased matrix effects were seen with the more specific methods. Both the Sirocco PPT and Ostro PPT with PL removal prepared samples had acceptable recoveries but substantial matrix effects (>25%), while the SLE prepared samples had the least recovery of all techniques. It should be noted that minimal optimization was performed for this or any technique, as one of our goals was to follow the recommended protocols for all products. It is possible that this performance would improve with additional protocol optimization. For the SPE techniques, this pattern of improved performance is more evident. All SPE techniques had adequate recoveries (>80%), with matrix effects decreasing from -40% for Oasis HLB to -13.6% for HLB PRiME and were negligible with Oasis MCX at 2.4%. Oasis MCX gave superior performance vs. WAX during screening with the Sorbent Selection Plate, so that sorbent was used for subsequent quantitative work. The other mixed-mode solvents screened, WCX and MAX had negligible recovery and are not shown in the Figure. It should be noted that the eluate from the first elution from the Sorbent Selection Plate was used for the mixed mode sorbents. Apixaban is not ionizable and is not expected to bind to the mixed mode sorbents via ion exchange, enabling us to elute in the methanol fraction alone while still leveraging the ion-exchange character of the mixed-mode sorbent to provide additional clean-up vs the RP-SPE techniques. As mentioned above, this enabled the screening of all 4 mixed-mode Oasis sorbents to rapidly determine the optimal mixed-mode SPE sorbent for this analysis.
Residual Phospholipids
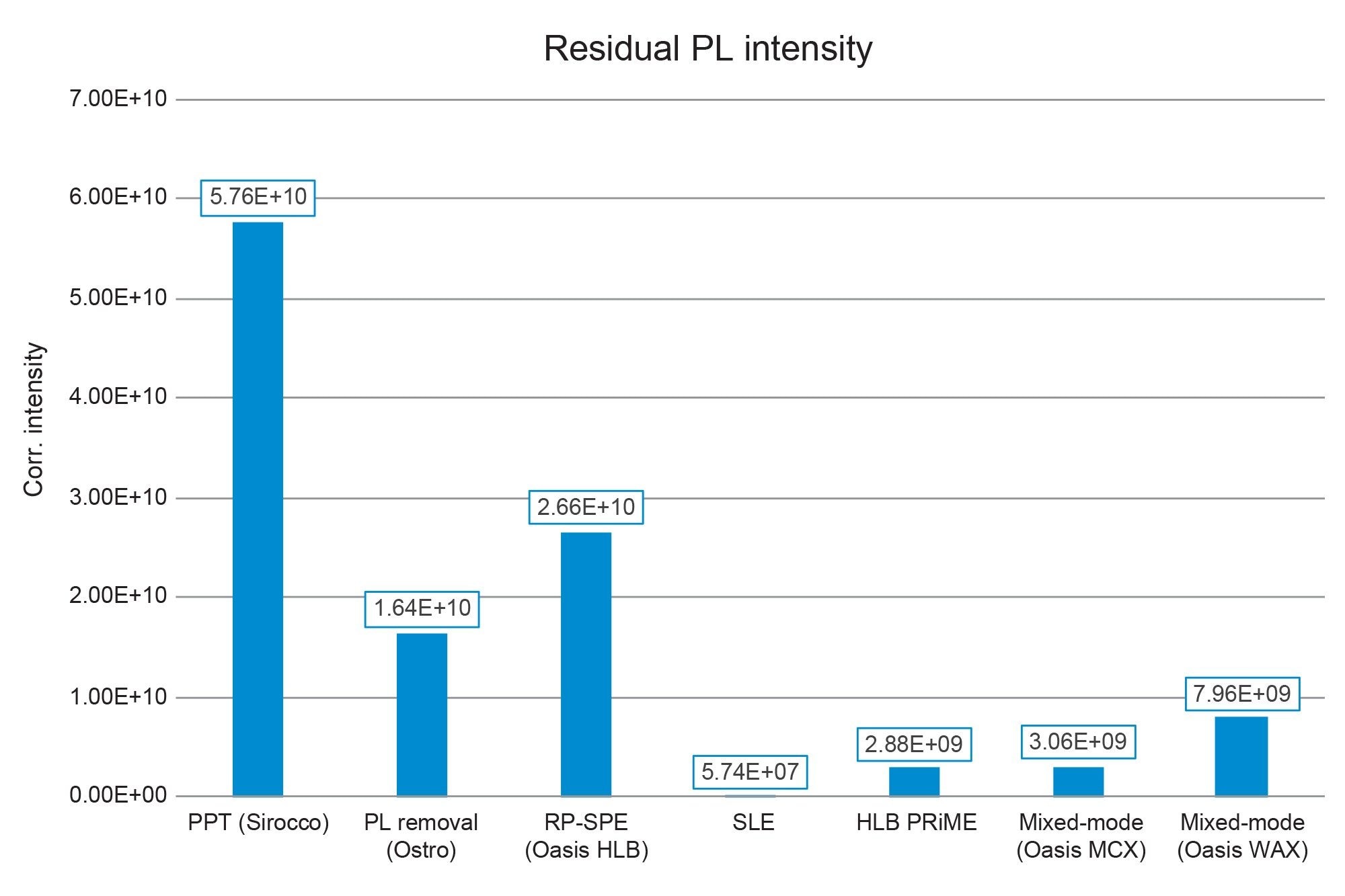
Residual phospholipids (PLs) were monitored as described in the Materials and Methods section. The absolute MS intensities of residual phospholipid traces were corrected for sample concentration or dilution and plotted in Figure 7. Unsurprisingly, simple PPT with Sirocco plates resulted in the highest presence of PLs. Use of the Ostro PPT plates significantly reduced the PL content of plasma extracts as expected. SLE prepared samples had negligible residual PLs, which has been seen before. [720006516]3 Looking at traditional SPE procedures, reversed phase SPE with Oasis HLB had the largest concentration of PLs, although they were reduced compared to PPT. Oasis HLB PRiME and Oasis MCX extraction reduced residual PLs by nearly 90% compared to standard reversed-phase SPE. These data provide additional information about the quality and cleanliness of the procedure. Combining this data with the recovery and matrix effects data in Figure 7 gives us a comprehensive view of the quality of the sample extraction procedures. In this case, we see that using Oasis MCX results in the best recovery with minimal matrix effects and low levels of residual PLs.
Quantitative Results
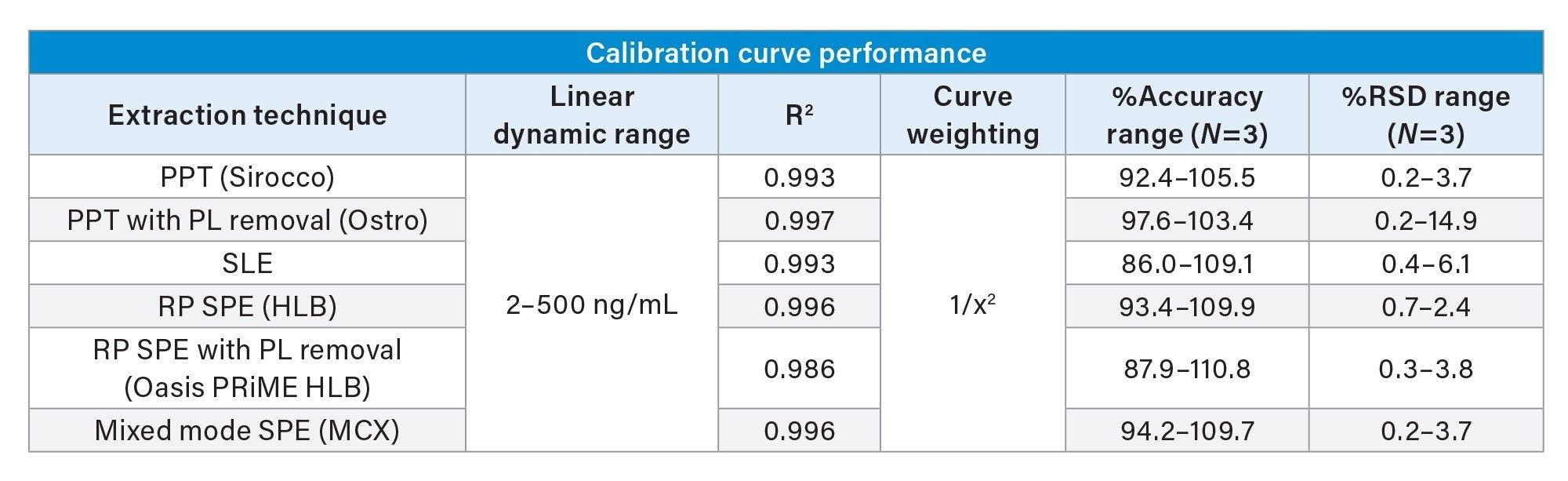
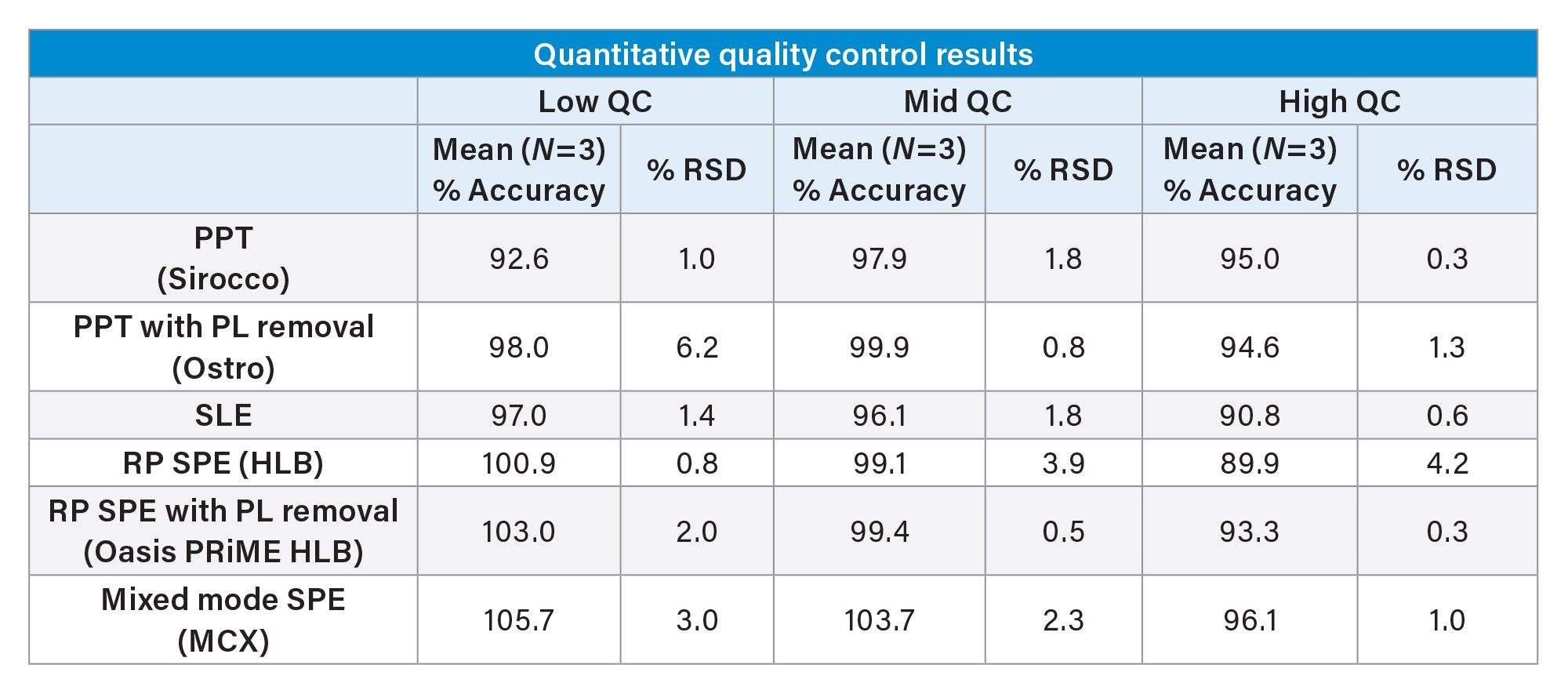
While the qualitative data in Figures 6 and 7 provide valuable insight into the choice of sample preparation method, the ultimate goal for bioanalytical methods is to achieve accurate, precise, and consistent quantitative data. Tables 2 and 3 below summarize the quantitative results from the automated extraction of standards and QC plasma samples using the sample preparation techniques described above. Calibration curves ranged from 2–500 ng/mL for all sample preparation techniques. Table 2 shows that calibrator accuracy ranges from 86–111% with RSDs <15% (N=3), with many in the single digits. This easily meets recommended bioanalytical method validation guidance criteria. A summary of the quantitative results associated with each technique is shown in Table 3. Although there were significant differences in recovery and matrix effects from the different techniques (Figure 6), quantitative performance was excellent for all extraction techniques, as is shown in Table 3, with mean QC accuracies all within 10% of nominal values. Precision was excellent as well with %RSD values in the single digits, and all but one were under 5%.
Conclusion
This application highlights a simplified bioanalytical extraction strategy, requiring no method development for successful extraction of apixaban from plasma, achieving high analyte recovery >75% with excellent reproducibility. Quantitative performance across the extraction techniques was excellent, with linear dynamic ranges from 2–500 ng/mL, QC accuracies ≥90% and RSDs ≤10% for apixaban extracted from plasma. The highest recoveries, lowest matrix effects and low residual phospholipids were obtained when doing more specific sample prep, in this case Oasis MCX. Other techniques had trade-offs in cleanliness and/or recovery. This application shows that even what is considered a more complicated sample preparation technique can yield outstanding results using a generic method with no additional method development. The combination of generic protocols and automated sample preparation with the Andrew+ Pipetting Robot configured with the Extraction+ Connected Device greatly simplified and streamlined sample extraction, and maximized lab productivity, reduced errors, and ensured overall analytical method performance.
References
- Zhang, X., Danaceau, J., and Chambers, E. Improvements in Recover, Reproducibility, and Matrix Effects with Oasis PRiME HLB, a Novel Solid-Phase Extraction Sorbent, Waters Application note. 720005495, September 2015.
- Danaceau, J., Wood, M. and Calton, L. Simultaneous Analysis of Diuretics and Beta-Blockers by Mixed Mode SPE and UPLC-MS/NS for Anti-Doping Analysis, Waters Application note. 720006515, March 2019.
- Danaceau, J., Haynes, K., and Chambers, E. A Comprehensive Comparison of Solid Phase Extraction (SPE) vs. Solid Liquid Extraction (SLE) vs. Liquid Liquid Extraction (LLE) Sample Prep Techniques in Bioanalysis and Forensic Toxicology Analyses, Waters Application note. 720006060, August 2017.
Featured Products
720007946, July 2023