For research use only. Not for use in diagnostic procedures.
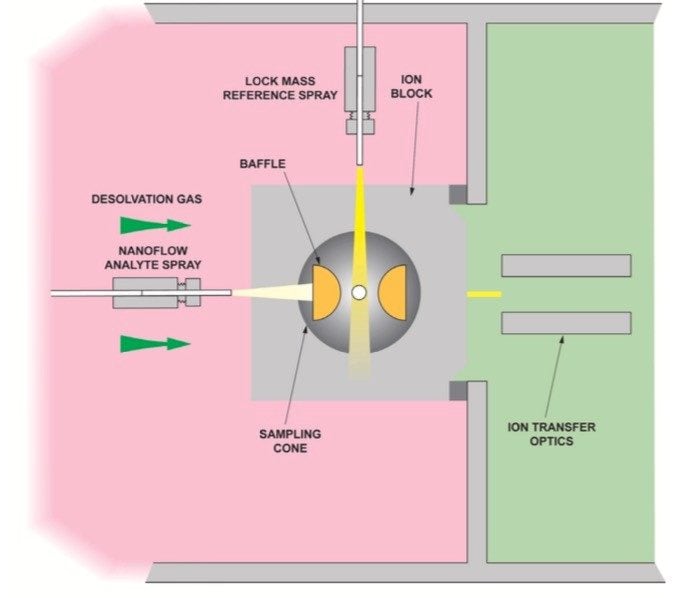
Due to the complexity encountered in the analysis of proteins obtained from mammalian systems, the primary route for the identification and characterization of the constituent proteins is electrospray (ESI) LC-MS/MS. The low endogenous levels and large dynamic range of proteins present in these samples dictate that nanoscale LC-MS/MS is often the method of choice, due to the concentration dependent nature of the ESI technique. This has led to nanoscale LC-MS/MS on a hybrid quadrupole orthogonal acceleration time-of-flight (Q-Tof) mass spectrometer as an established technique for high sensitivity identification and characterization of proteins. Typically, these experiments employ HPLC columns that have an internal diameter of 75 μm, or less, operating at flow rates of approximately 200 nL/min. While this set-up offers the optimum sensitivity it does not allow the post-column addition of an internal reference ion, as this would detrimentally effect the resolution of the HPLC separation, resulting in peak broadening. The use of an internal reference is required to provide reliable high mass measurement accuracy. Here we report the use of a nanoflow LockSpray interface to routinely provide enhanced mass measurement in the analysis of protein digests.

Initial results were obtained from the LC-MS/MS analysis of 500 fmoles of a tryptic digest of Bovine Serum Albumin. Data were acquired automatically from a Q-Tof micro fitted with NanoLockSpray, using DDA.
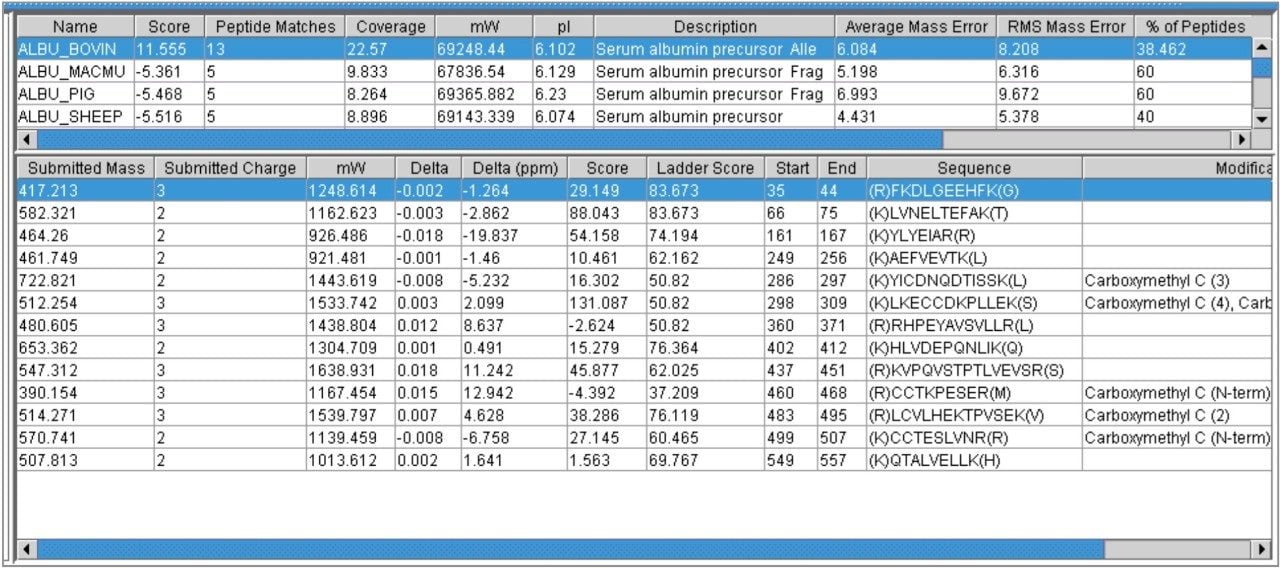
Results from this analysis are presented in Figure 3, where the databank search was conducted with a precursor ion tolerance of 20 ppm. In this case 13 matching peptides were identified to the BSA sequence. Several other serum albumin proteins from different species were also identified. The RMS error for the peptides matching to the BSA sequence was 8 ppm.
Analysis of the same data set using a 100 ppm window resulted in the identification of BSA as the top hit, with again 13 matching peptides, however an incorrect identification Ribonuclease R was also returned (Figure 4).
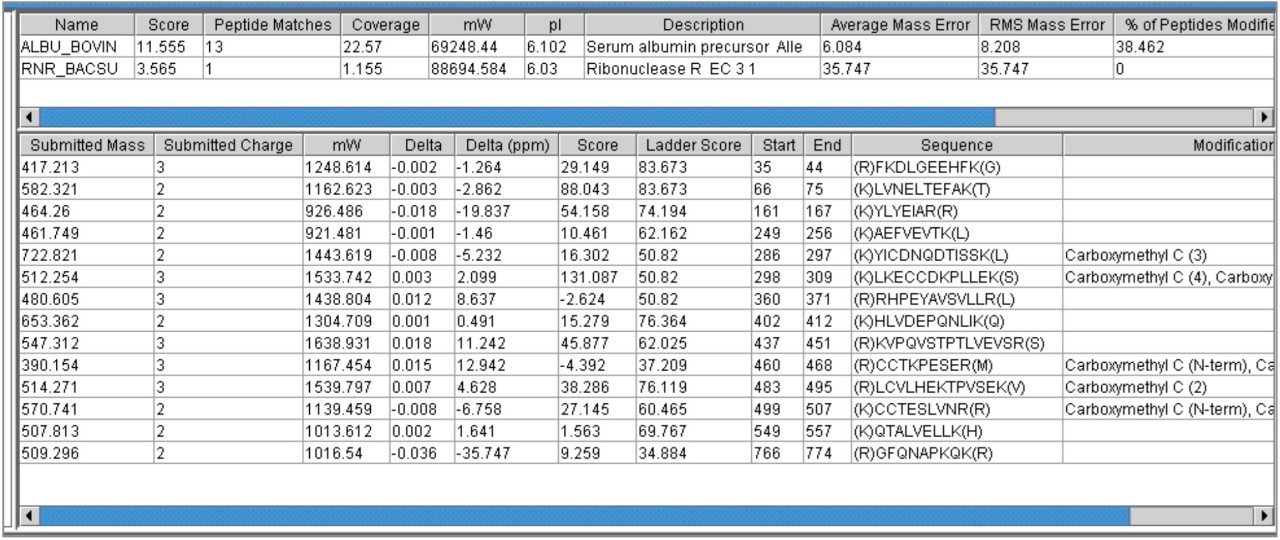
Despite the poor quality of this identification, it would require manual verification to remove it from the protein hit list.
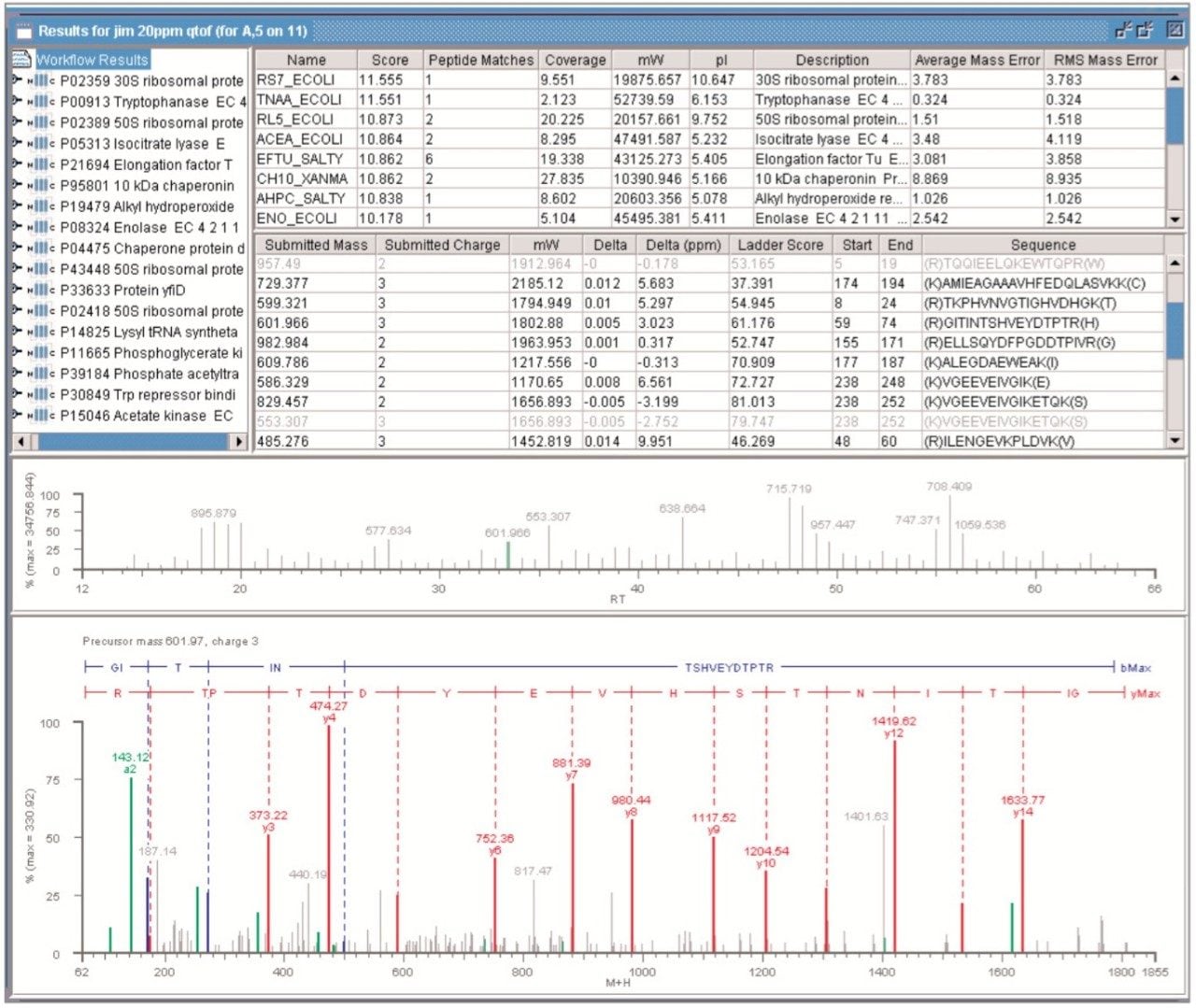
Figure 5 shows the results from a DDA analysis of an Escheria coli (E. coli) sample containing multiple proteins. In this case the SWISS-PROT databank was searched with the peptide precursor ion tolerance set to 20 ppm.
All of the retrieved proteins originated from E. coli. Identification of 17 proteins was made. The peptides matching the parent proteins had an RMS error of 10 ppm or better.
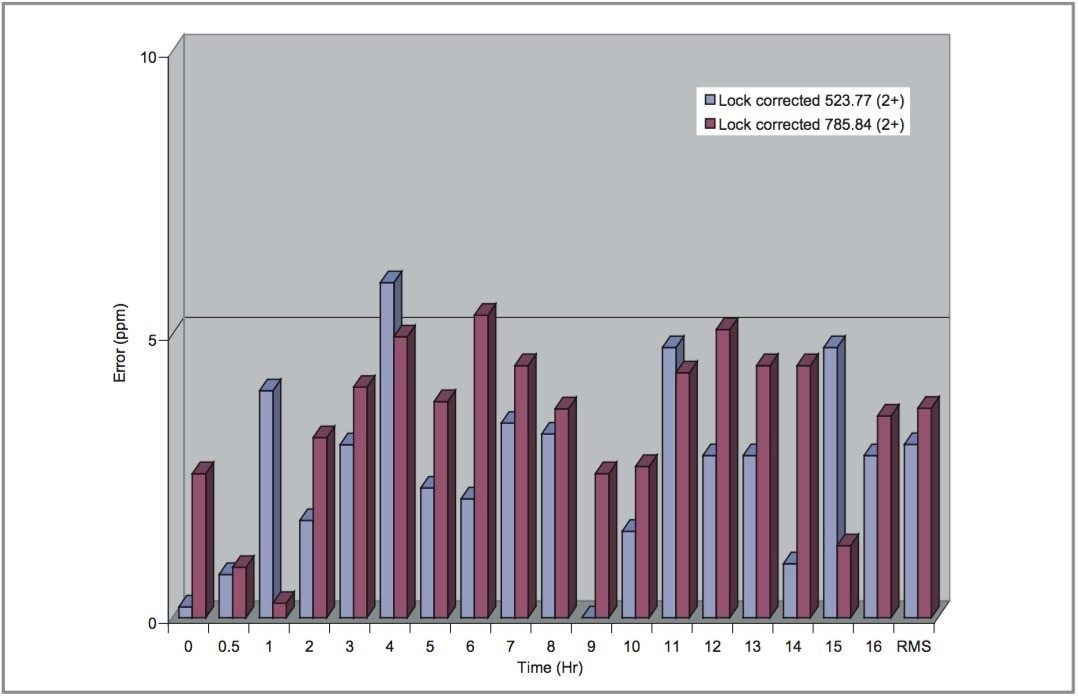
Stability of mass measurement obtained with the NanoLockSpray source, over an extended period of time, was investigated through the infusion of two peptides, [Glu’]-fibrinopeptide B (m/z 785.8426 2+) and Angiotensin II (m/z 523.7751, 2+) over a 16-hour period (Figure 6). Mass measurement errors obtained from the Q-Tof micro equipped with the NanoLockSpray source were determined with the external lockmass correction. It can be seen that errors, when the single point external calibration was used, were 5 ppm or better over the 16-hour period.
During a typical nanoflow LC-MS/MS experiment the tryptic peptides present exhibit a wide dynamic range and as such are detected by the mass spectrometer with varying signal intensities. This provides a challenge to achieving routine exact mass measurement as very intense peaks can cause the detector of the mass spectrometer to become saturated or move into 'dead time'.
Once a single ion has been detected there is a period during which further ion arrival events will not be detected. The result is a non-linear response between ions detected vs ions arriving at the detector. This manifests itself in a shift to a lower mass being reported for that particular ion.
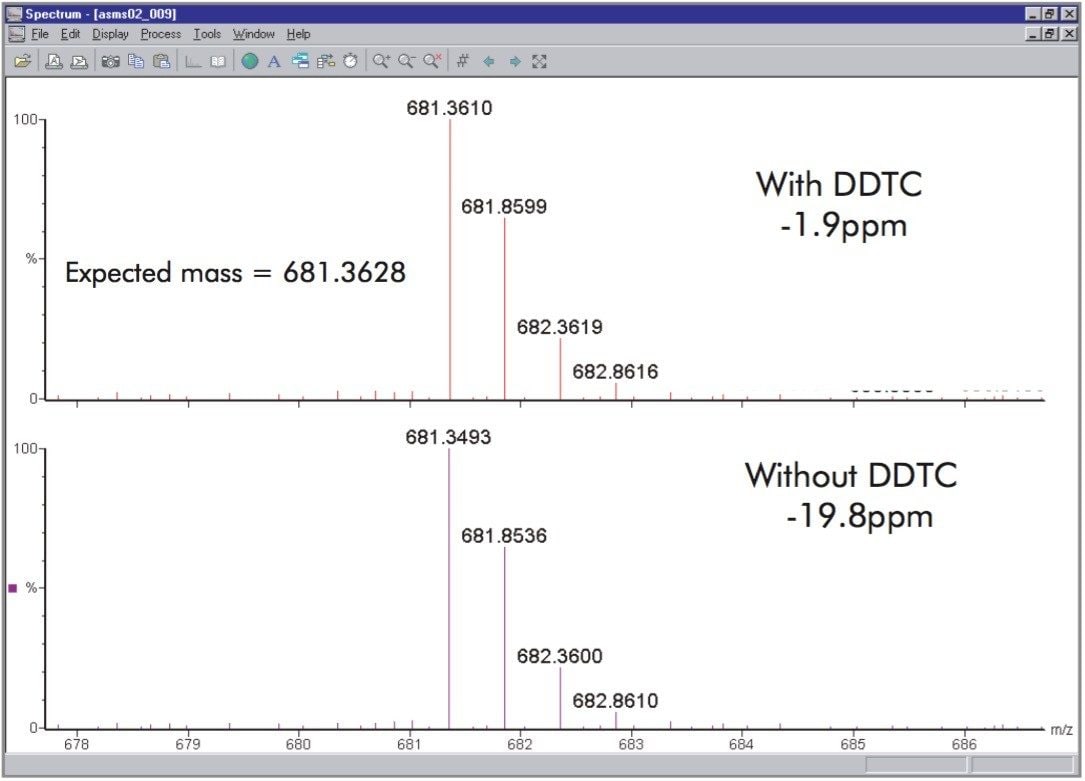
An algorithm termed Digital Dead Time Correction (DDTC) can be used to correct for this effect. An example of this effect is presented for data obtained from a Q-Tof Ultima API. This is presented in Figure 7 where a doubly charged ion at m/z 681.36 is shown both with and without DDTC applied during processing. The ion was assigned the sequence IQTQPGYANTLR during database searching. With DDTC applied the mass measurement for the ion was -1.9 ppm and without DDTC applied it was measured to be -19.8 ppm.
A challenge remains in an LC-MS/MS experiment in that often many of the MS/MS spectra acquired do not provide matches when searched against known protein sequence databanks. The nature of database searching is such that only peptides that match exactly those within the databank will be identified. Consequently, many good quality spectra of novel peptides or of those containing a single amino acid substitution, or modification remain unmatched. Similarly, samples from organisms poorly represented in sequence databases can often produce a large number of unidentified spectra. Currently a common solution to this would be to extract these spectra manually from the data set, derive some degree of peptide sequence and perform further database searching. This, however, can be time consuming when large numbers of spectra are involved and is reliant upon the skill of the operator.
With the introduction of an automated computer-sequencing algorithm, MS/MS spectra can be identified by direct derivation of the novel peptide sequence through a double application of a Bayesian probabilistic analysis. A fragmentation model is applied to compare trial sequences against singly charged, de-isotoped MS/MS spectra. An overall confidence value for the most probable sequence, along with a confidence in the assignment of each individual residue, is calculated.
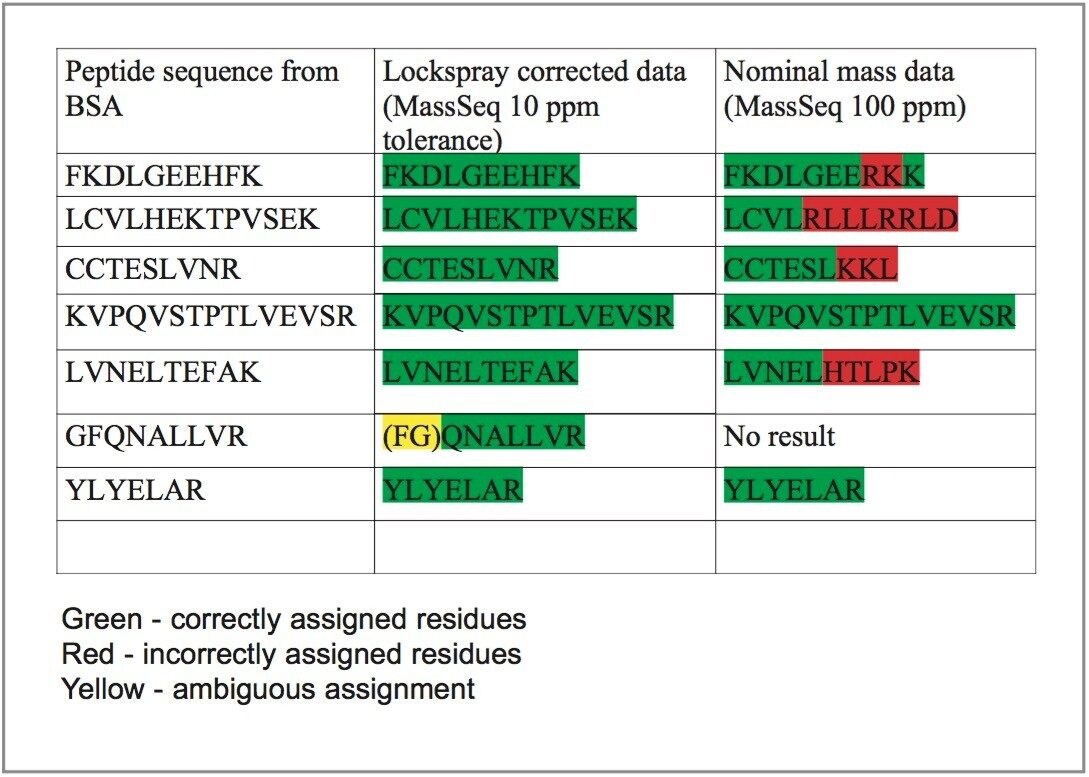
The MassSeq algorithm (ProteinLynx Global SERVER v2.0) utilizes the exact mass measurement of the MS/MS fragment ions to confidently define the most probable amino acid sequence. This is illustrated in Figure 8 where MS/MS spectra were acquired at different mass measurement accuracy using an ESI Q-Tof. It can be seen, in the resulting sequence determinations, that the data measured to less than 10 ppm provides significantly better results than that acquired at 100 ppm.
720000738, September 2003