
A quick and simple procedure is presented for the extraction of aminoglycoside antibiotics from bovine meat and milk. A single operator can easily analyze 20 samples per day.
Aminoglycoside antibiotics, used in veterinary medicine for the treatment of animals bred for meat and milk production, require an effective method for residue analysis in these commodities. This class of antibiotics presents significant challenges for residue analysis. Unlike most other antibiotic classes, these compounds are not amenable to extraction from tissue or milk using acetonitrile or other organic solvents. In this study, the aminoglycosides are extracted from meat tissue or milk using an aqueous buffer with trichloroacetic acid (TCA) added to precipitate protein and inhibit protein binding of the analytes. An effective SPE-based cleanup procedure is employed prior to LC-MS analysis to remove the residual TCA and to minimize co-extracted interferences. For SPE, good recovery and cleanup were obtained for bovine milk and meat using Oasis HLB, a high-performance, water-wettable, reversed-phase sorbent. In the subsequent analysis, excellent UPLC performance was obtained using the ACQUITY UPLC HSS PFP Column in reversed-phase ion-pairing mode. Heptafluorobutyric acid (HFBA) was used as the ion-pairing reagent. This reagent is volatile and compatible with mass spectrometry.
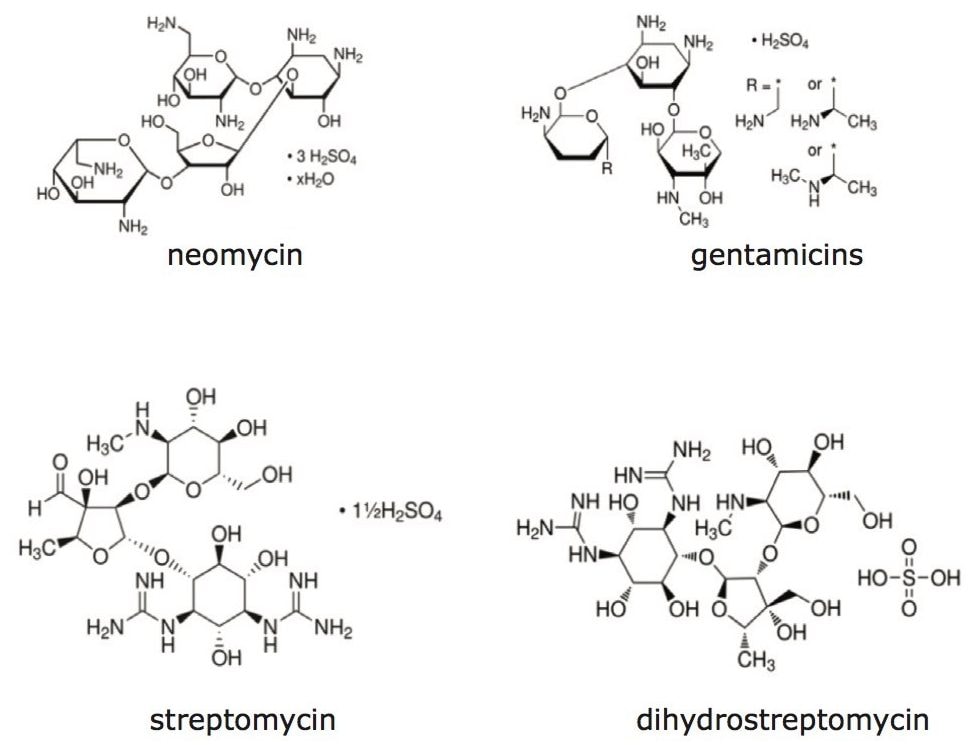
Worldwide maximum residue levels (MRLs) for the aminoglysoside antibiotics generally range from 100 to 1000 µg/kg milk, as shown in Figure 1. This application note presents methodology suitable for ppb level determination of aminoglycoside antibiotics in bovine milk and tissue and was developed in part from methods presented in references 1 and 2.
|
System: |
ACQUITY UPLC |
|
Column: |
ACQUITY HSS PFP, 1.7 μm, 2.1 x 100 mm |
|
Injection volume: |
30 μL |
|
Temp.: |
35 °C |
|
Mobile phase A: |
20 mM HFBA in water |
|
Mobile phase B: |
20 mM HFBA in acetonitrile |
|
Flow rate: |
0.50 mL/min |
|
Gradient: |
20% B initial, linear gradient to 80% B in 7 min, hold for 8 min, back to 20% B for 8.1 min. Hold and re-equilibrate for 10 min. |
|
Mass spectrometer: |
ACQUITY TQD |
|
Mode: |
Positive electrospray (ES+) |
|
Capillary: |
3.0 kV |
|
Extractor: |
3.0 V |
|
Source temp.: |
130 °C |
|
Cone gas: |
20 L/h |
|
Desolvation temp.: |
450 °C |
|
Desolvation gas: |
900 L/h |
|
Collison gas: |
Argon at 0.20 mL/min |
Extraction Buffer (10 mM NH4OOCH3/0.4 mM Na2EDTA/1% NaCl/2% TCA): Place 0.77 g of ammonium acetate (NH4OOCH3) into a 1-L volumetric flask. Add approximately 900 mL of reagent water and dissolve. Adjust pH to 4.0 with 1 N HCl or 1 N NaOH. Add 0.15 g disodium ethylenediamine tetraacetate (Na2EDTA.2H2O), 5 g of sodium chloride (NaCl), and 20 g of trichloroacetic acid (TCA). Mix well to dissolve and bring to the mark with reagent water.
Place 2 g homogenized bovine tissue or 10 mL milk into a 50 mL centrifuge tube. Add 20 mL extraction buffer, vortex for 10 s, then shake well for 1 min. Centrifuge the sample at 4000 RPM for 5 min, and collect the supernatant. Adjust the pH of the supernatant to 6.5±0.5 using diluted HCl or NaOH as needed.
An Oasis HLB 96-well Plate (30 mg) was used in this study. A 1-cc, 30-mg cartridge can be used if desired. Condition the well or cartridge with 1.5 mL methanol, followed by 1.5 mL water. Set the flow rate at 1 mL/min or less. Load the pH-adjusted supernatant obtained from the initial extraction; a 1-mL aliquot is loaded for tissue samples, a 1.5-mL aliquot for milk samples. Wash with 1 mL water. Elute with 0.5 mL 10:5:85 formic acid/isopropanol/water. Add 1.5 µL HFBA and analyze using UPLC-MS/MS.
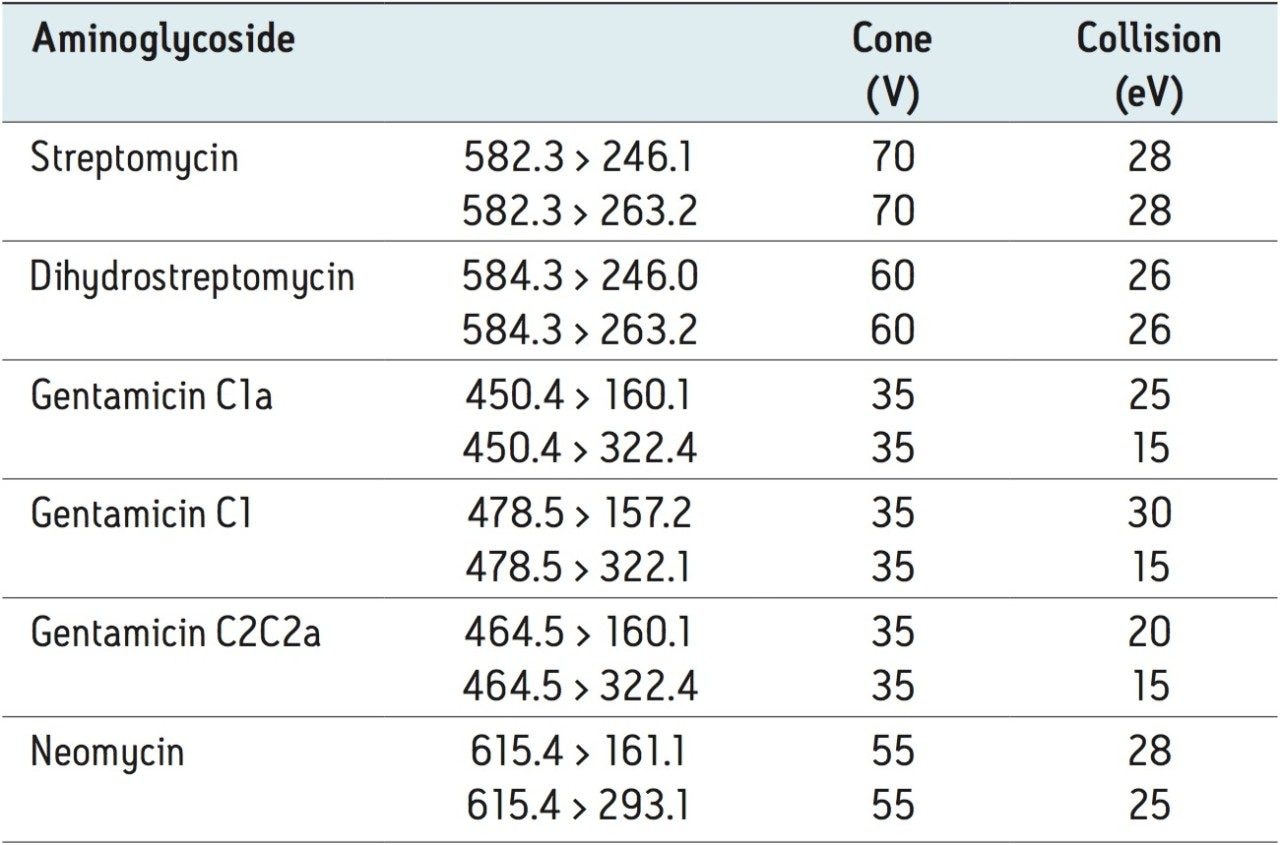
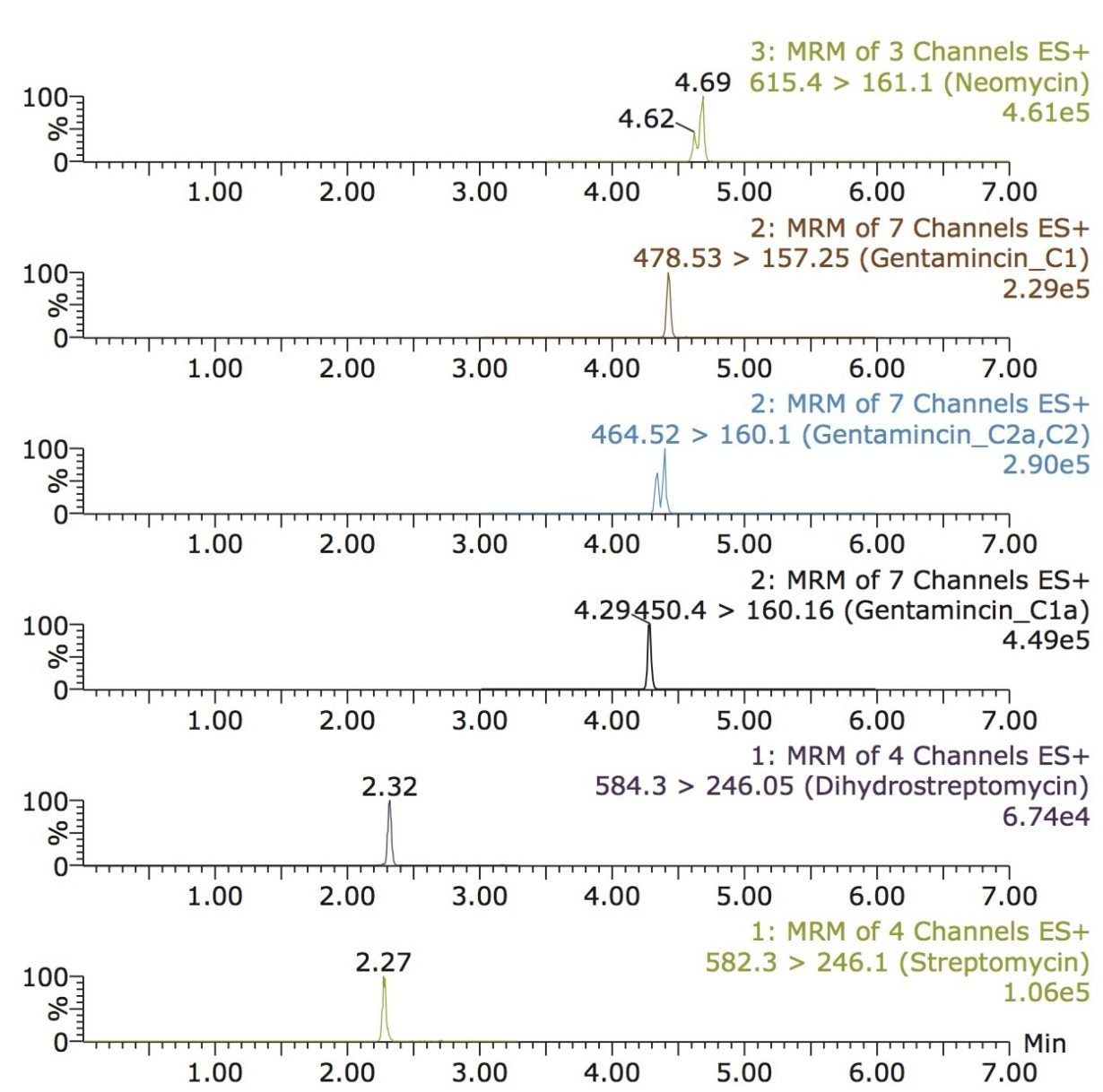
Figure 2 shows a reconstructed UPLC-MS/MS chromatogram obtained from the analysis of a spiked bovine milk sample. The same conditions were used for the bovine muscle samples. The results obtained from the spiking experiments are shown in Table 2 (milk) and Table 3 (muscle tissue). Neomycin (B and C) and gentamycin (C2a and C2) exist as a mixture of isobaric isomers and are partially resolved. The peak area for these pairs was summed for quantification.
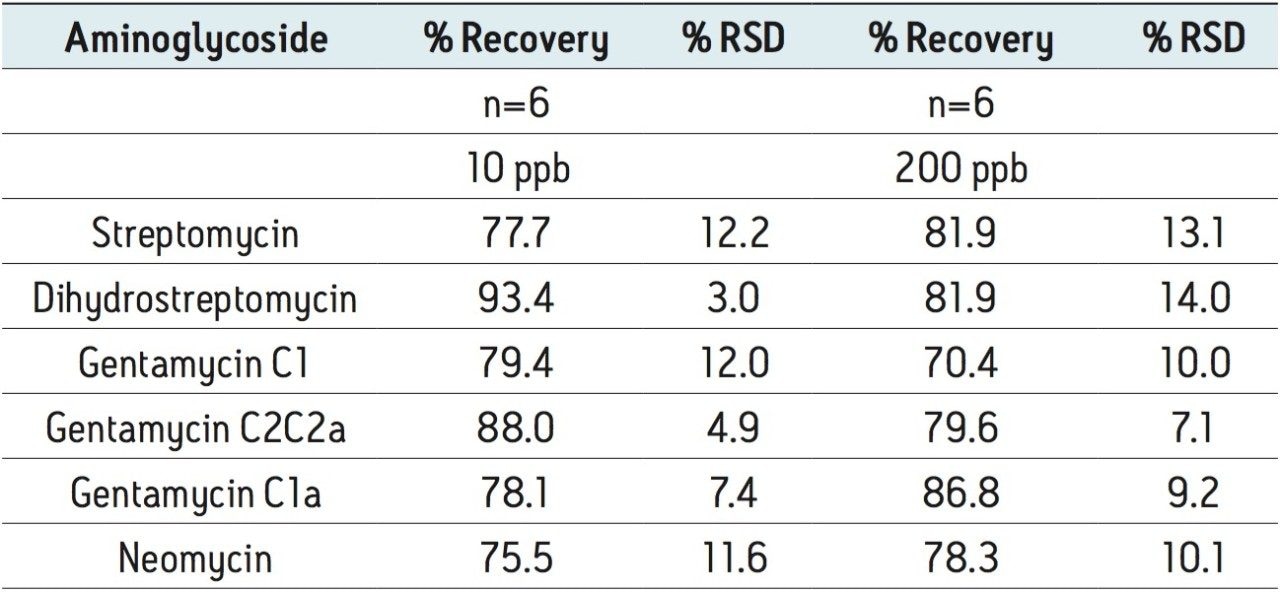
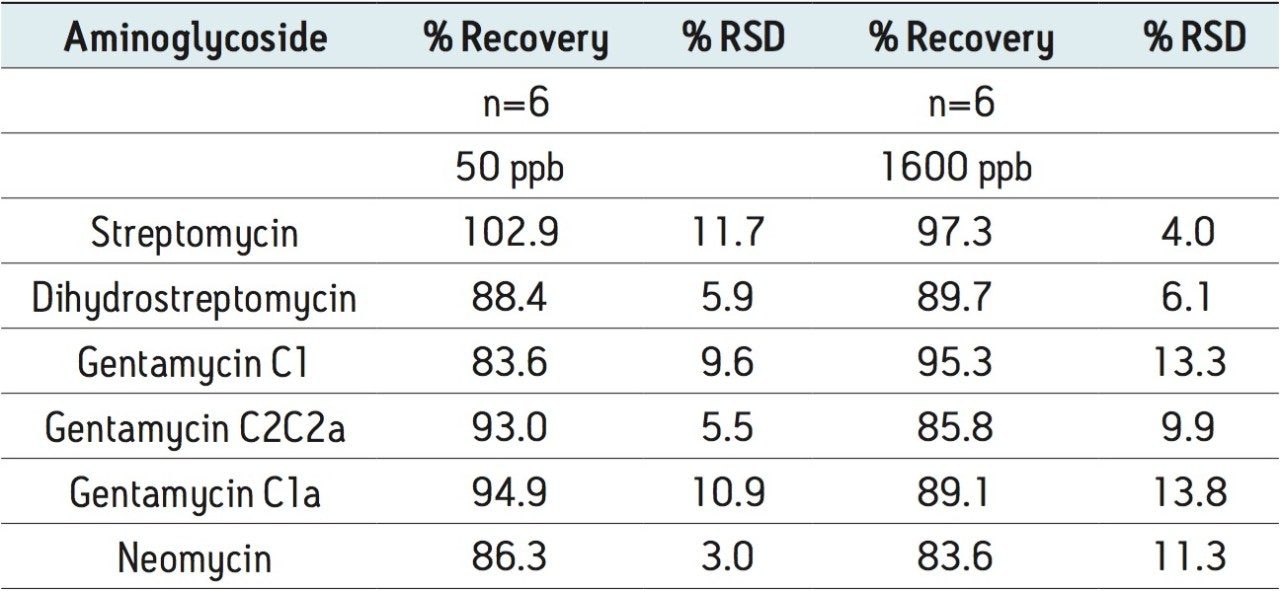
Recovery was determined by comparing the MRM peak areas for samples spiked into the sample matrix prior to sample preparation with the peak areas for samples spiked after all sample preparation steps.
Matrix effects were less than 30% for meat and less than 12% for milk.
Unlike many other antibiotics used in veterinary medicine, the aminoglycosides cannot be extracted from animal tissues and related samples using organic solvents. However, these compounds can be effectively extracted from tissue with an aqueous buffer. This extraction buffer also includes an agent (TCA) to facilitate protein precipitation.
The use of a low organic content eluent for the Oasis HLB SPE protocol gave improved cleanup compared with an alternative methanol-based elution (samples eluted with formic acid/methanol had iomatrix effects greater than 50%). Apparently, significant matrix interference components remain on the sorbent after elution of the aminoglycosides with the aqueous eluent.
The ion-pairing reversed-phase mode was chosen for the UPLC analysis. The best compromise for peak shape and sensitivity was obtained using 20 mM heptafluorobutyric acid as the ion-pairing agent. UPLC using HILIC columns was also considered; however, peak shape was not as sharp as the chosen method. Also, a disadvantage of the HILIC mode is that the diluent for standards and samples should be acetonitrile. The optimized SPE protocol presented in this application note provides an aqueous-based sample for injection that is better suited for reversed-phase analysis.
720004512, December 2012