For research use only. Not for use in diagnostic procedures.
Biomarker discovery and validation are the first two steps in understanding disease and drug development. This application note has demonstrated the utility of the novel ionKey/MS System for rapid and robust discovery validation experiments. Quantification measurements for light to heavy stable isotope labeled peptides have shown excellent consistency and are in agreement with expected values.
The ionKey/MS System is a robust, novel microfluidics platform that eliminates manual chromatographic connections and enables high throughput and robust validation analysis.
Biomarker discovery and validation are the first two steps in understanding disease and drug development. Validation is technology-challenged since it requires analyzing a large number of samples with high-throughput, but nevertheless requires high sensitivity, high resolution, large dynamic range and excellent selectivity.
Targeted LC-MS based assays afford protein quantification with the reproducibility and throughput required to improve marker acceptance. Multiple reaction monitoring (MRM) using tandem quadrupole mass spectrometers is one of the enabling technologies applied in targeted LC-MS approaches.1 Overall, MRM-based methods compare favorably with antibody-based techniques, such as ELISAs or protein arrays, in that MRM-based methods are less expensive and can be developed more rapidly. However, one of the major challenges using MRM for candidate marker verification in mammalian body fluids is the required sensitivity for the quantification of low-abundance proteins, especially in the case of sample limited conditions.
Miniaturized LC systems offer improved mass sensitivity but often lack the required throughput, robustness, and reproducibility. The application of a novel microfluidics platform for the quantification of marker peptides and proteins is presented, considering speed, sensitivity, and selectivity.
|
LC system: |
nanoACQUITY UPLC System or ACQUITY UPLC M-Class System |
|
Sample loop: |
5 μL |
|
Separation device: |
iKey BEH C18 Separation Device, 130Å, 1.7 μm, 150 μm x 100 mm (p/n 186007258) |
|
iKey temp.: |
40 °C |
|
Flow rate: |
1.2 μL/min |
|
Mobile phase A: |
98.9:1:0.1% v/v water/acetonitrile/ formic acid |
|
Mobile phase B: |
98.9:1:0.1% v/v acetonitrile/water/ formic acid in water |
|
Volume injected: |
0.1 to 1.0 μL |
|
Time(min) |
%A |
%B |
Curve |
|---|---|---|---|
|
Initial |
98 |
2 |
Initial |
|
1.0 |
98 |
2 |
6 |
|
45 |
60 |
40 |
6 |
|
46 |
15 |
85 |
6 |
|
47 |
15 |
85 |
6 |
|
48 |
98 |
2 |
6 |
|
MS system: |
Xevo TQ-S |
|
Acquisition mode: |
MRM |
|
Quadrupole resolution: |
0.4 Da or 0.7 Da |
|
Ionization mode: |
ESI positive |
|
Capillary voltage: |
3.0 kV |
|
Source temp.: |
100 °C |
MassLynx raw data were analyzed using Skyline2 and visualized using Spotfire DecisionSite (Tibco Spotfire, Boston, MA).
MS Qual/Quant QC Mix was obtained from Sigma-Aldrich (St. Louis, MO, USA). MassPREP E. coli Digest Standard (p/n 186003196) was from Waters Corporation (Milford, MA, USA).
The MS Qual/Quant mixture was spiked into the E. coli background such that loads for a 1 μL injection ranged from 32 amol to 40 fmol peptides in the presence of 100 ng E. coli. The sample was injected three times at four different loadings (0.1, 0.2, 0.5, and 1 μL).
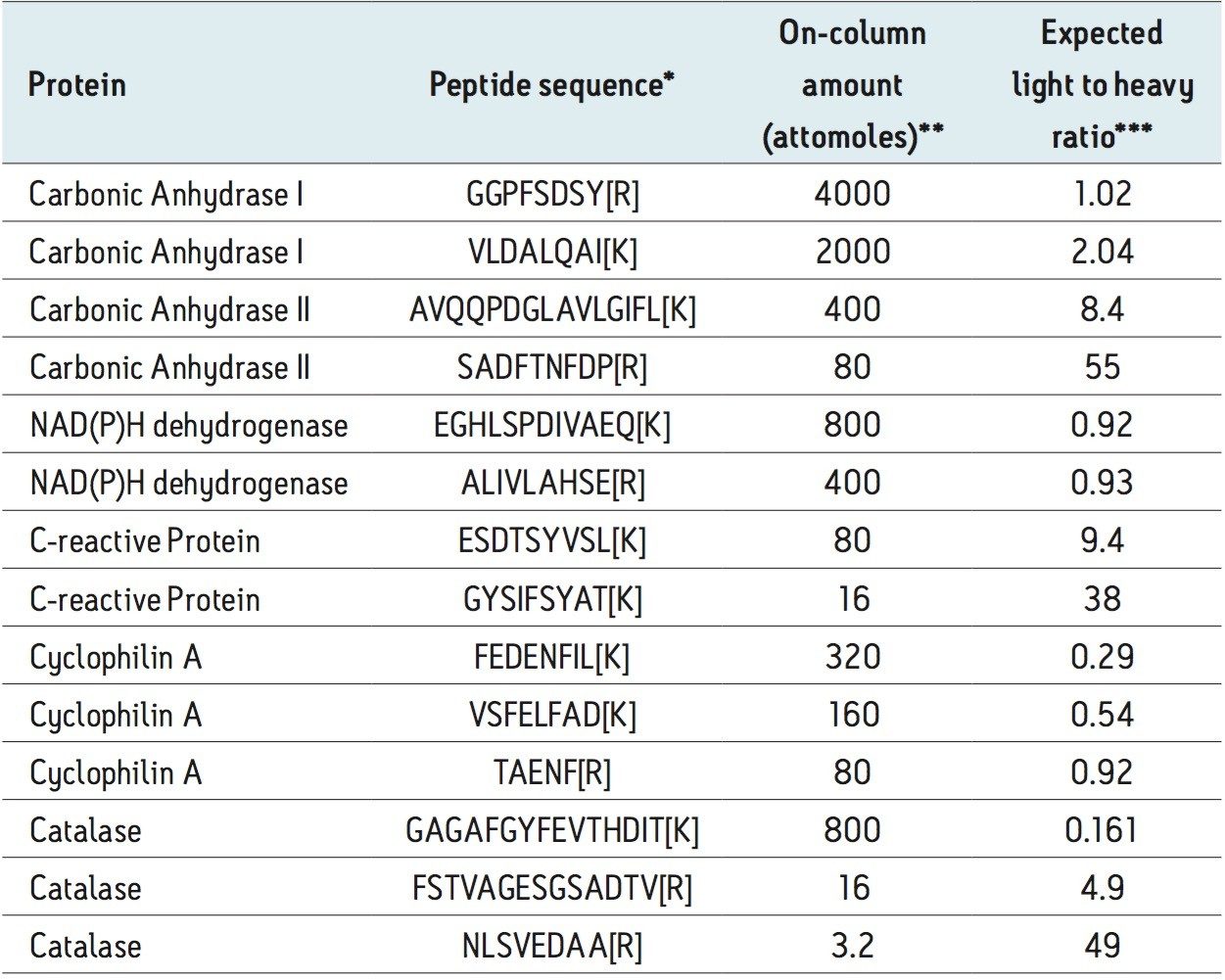
The MS Qual/Quant QC mixture consists of 14 peptide species present as light and heavy labeled analogues and at varying amounts to give an in-sample dynamic range of 1.25e3, Table 1. The peptide mixture was spiked into a complex background matrix of an E. coli tryptic digest to represent a high-throughput validation study and access quantitative precision and accuracy.
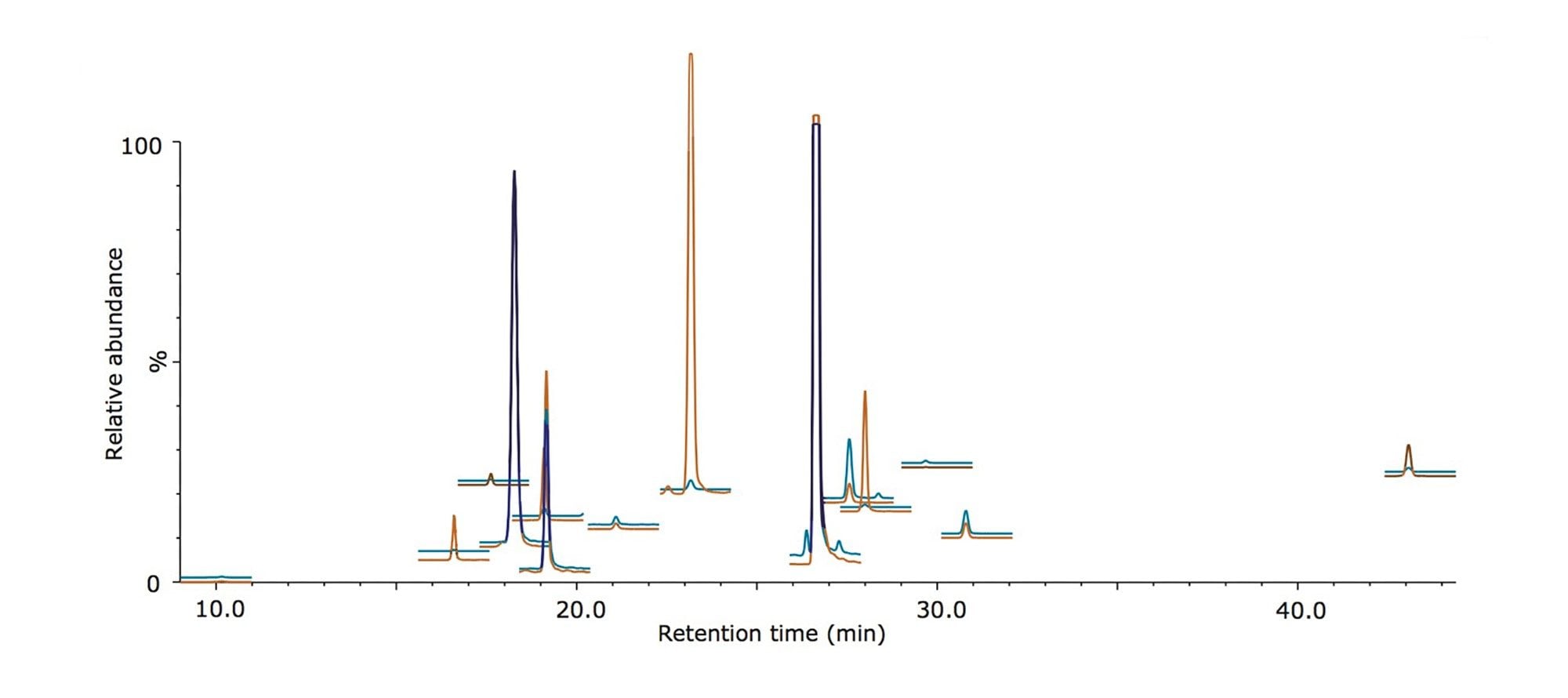
An MRM method containing 28 functions was programmed to monitor three transitions from each of the light and heavy peptide species, and the sample was analyzed using the ionKey/MS System with the nanoACQUITY UPLC System and the Xevo TQ-S Mass Spectrometer. Figure 1 shows the extracted MRM chromatograms for each peptide species resulting from a 0.5 μL injection and exhibits i) peptides that are resolved from the background matrix and ii) the dynamic range present within the sample. The on-column amounts displayed here range from 16 amol for the peptide NLSVEDAA[R] to 20 fmol for the peptide GGPFSDSYR.
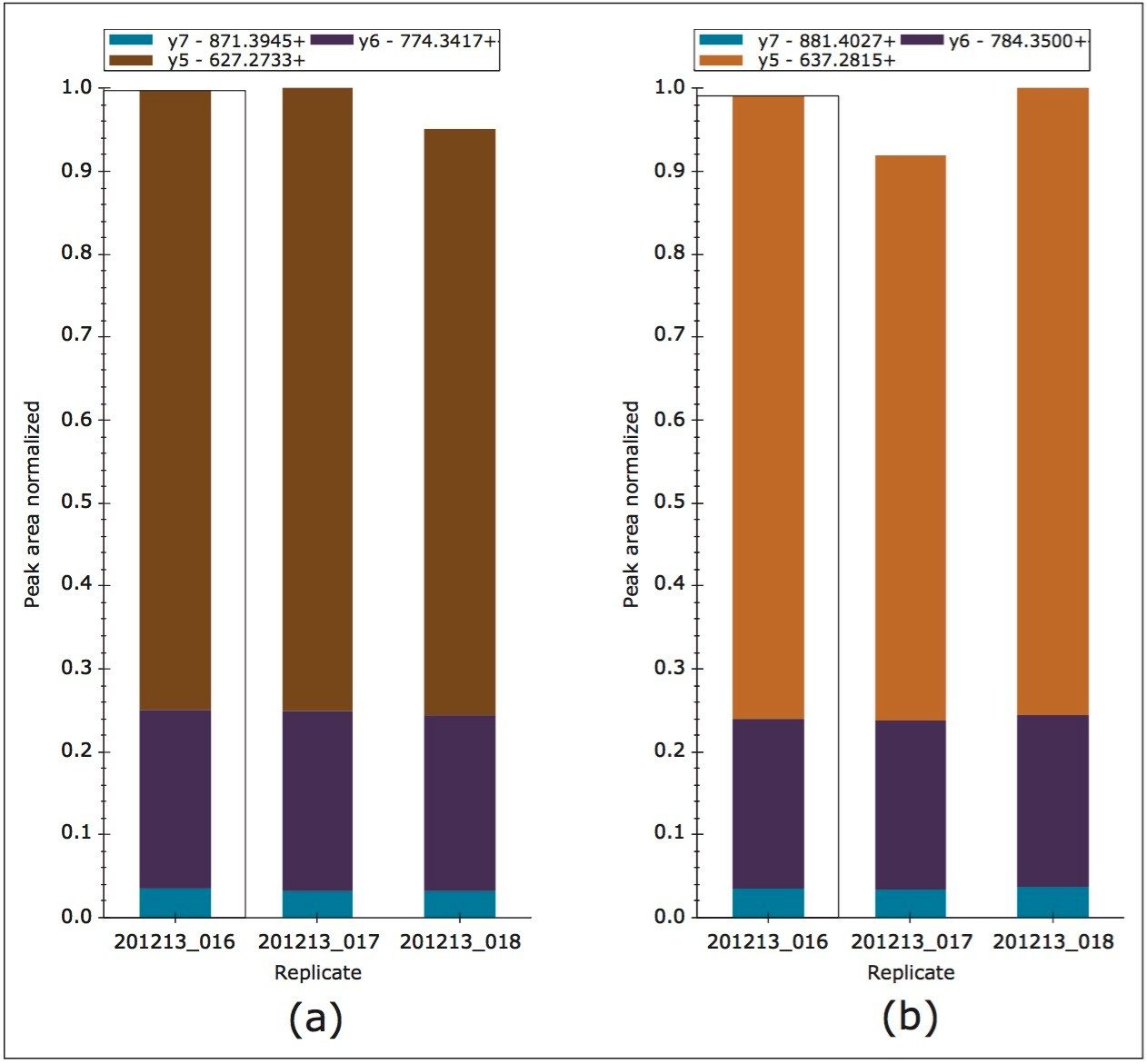
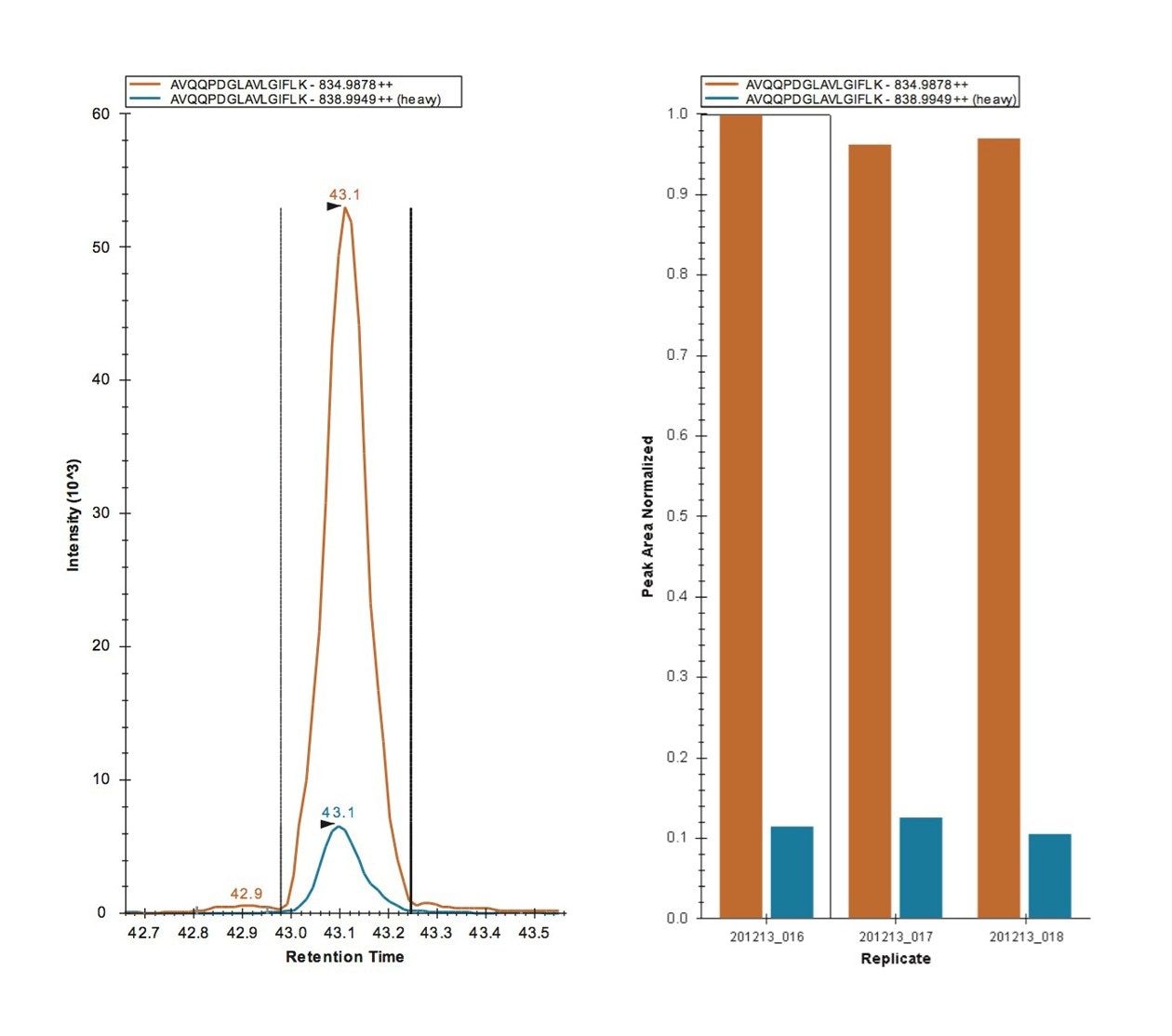
MRM transitions were inspected using Skyline, ensuring that a minimum of three peptides per protein and three transitions per peptide were detected and analyzed. Figure 2 represents the technical reproducibility of three transitions for an example light/heavy peptide, i.e. fragment ions y5, y6, and y7 for peptide GGPFSDSYR. Further Skyline interrogation shows that excellent quantitative measurement consistency, even without normalization, between technical replicates is readily achieved, Figure 3. The example illustrates mass chromatograms and ratio measurements for peptide AVQQPDGLAVLGIFLK from Carbonic Anhydrase II, which is present in the mixture at elevated levels for the light analogue.
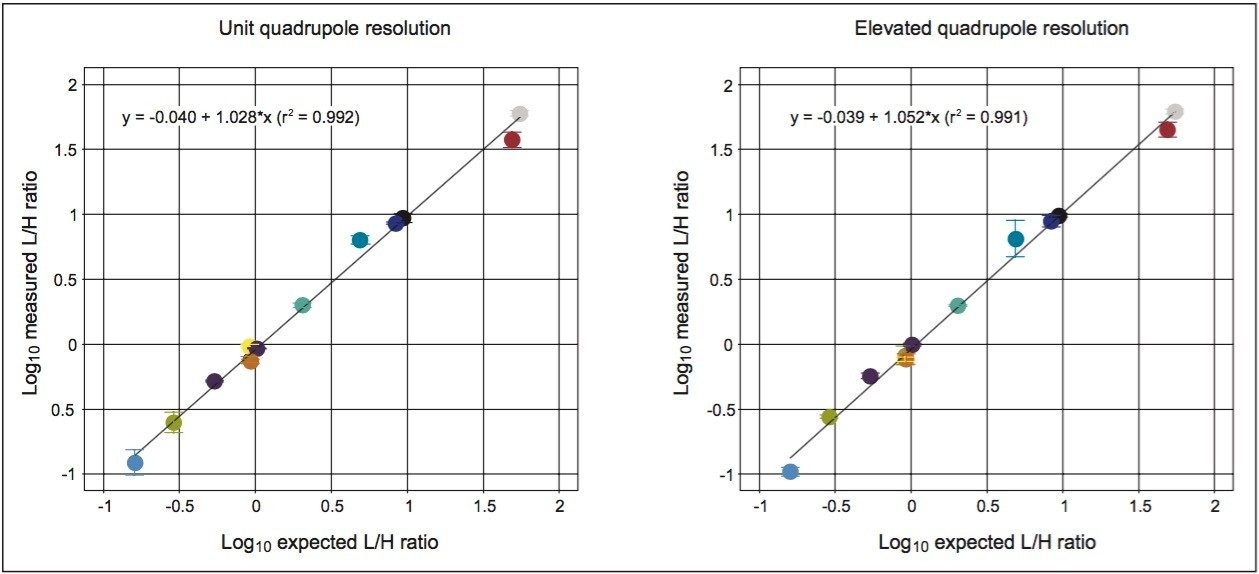
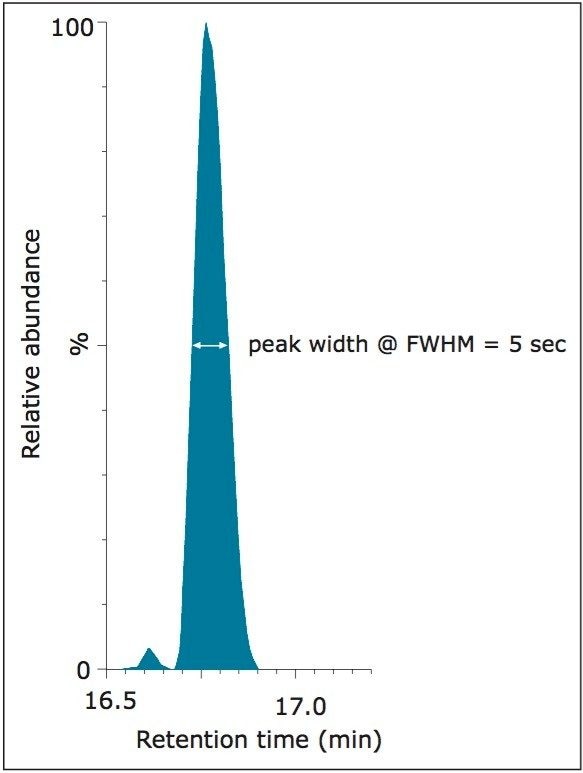
In Figure 4, the measured light-to-heavy ratios for every injection, 12 for each quadrupole setting, and every peptide in the mixture, are plotted against the expected ratios from the supplier certificate of analysis at unit and elevated quadrupole resolution settings. The results clearly show that excellent ratio measurements are achieved over the range of expected ratios for both mass spectrometer quadrupole resolution settings. For this application, unit quadrupole resolution afforded sufficient quantitative accuracy. The results presented are the average relative quantification results for four different injection volumes, ranging from 0.1 to 1 μL. This covers a sample load from 3.2 amol for NLSVEDAA[R] to 40 fmol for GGPFSDSY[R] and VLDALQAIK,spanning four orders of concentration dynamic range. As can be observed by the error measurement values, precision was not noticeably affected by quadrupole resolution. As an indication of the limits of detection that can be achieved, shown in Figure 5 is the chromatogram for the peptide NLSVEDAA[R] from Catalase. This chromatogram was generated from the lowest sample injection amount, 0.1 μL and so equates to 3.2 amol on-column.
This application note has demonstrated the utility of the novel ionKey/MS System for rapid and robust discovery validation experiments. Quantification measurements for light to heavy stable isotope labeled peptides have shown excellent consistency and are in agreement with expected values. The limit of detection has been demonstrated down to at least 3.2 amol on-column.
720005010, January 2016