
Previous work presented described the use of the Waters Screening Platform Solution in combination with Waters’ toxicology library to initially screen a local well water sample for the presence of a large number (>1000) of PPCPs, pesticides and drugs of abuse.2 In this application note, we have processed the same dataset with the metabolite identification aspect of the integrated software system to isolate known and potential metabolites of the confident screening matches in the dataset. Using the metabolite identification functionality of UNIFI, three metabolites of carbamazepine were identified with confidence in an enriched local well water sample.
In recent years, there has been increasing concern regarding the presence of pesticides, pharmaceuticals, and personal care products (PPCPs) in water bodies throughout the world.1 A greater demand is being placed on techniques not only used to screen for these compounds, but to screen for the presence of their metabolites.
Data obtained from a non-targeted acquisition on a high resolution mass spectrometer can be used to target a theoretical unlimited number of compounds. Moreover, information rich datasets collected using UPLC/MSE can be used to reduce the large number of false detects that arise when targeting a large number of compounds verses accurate mass as a sole point of contaminant identification. MSE provides accurate mass measurements for both precursor and fragment ion information in a single experiment by alternating scans between low and high collision energies. In combination with UNIFI, an integrated scientific information system, it is now possible to screen for the presence of PPCPs, their adducts, and potential metabolites in a routine laboratory environment.
Previous work presented described the use of the Waters Screening Platform Solution in combination with Waters’ toxicology library to initially screen a local well water sample for the presence of a large number (>1000) of PPCPs, pesticides and drugs of abuse.2 In this application note, we have processed the same dataset with the metabolite identification aspect of the integrated software system to isolate known and potential metabolites of the confident screening matches in the dataset. Once discovered, metabolites were made available for future screening experiments by adding the detection results (retention time and identified fragment ions) into a scientific library.
A locally obtained well water sample was enriched one thousand times as previously described.2,3 A comprehensive dataset, collected using UPLC-MSE was obtained within UNIFI. The toxicology screening solution within UNIFI contains pre-defined LC-MS conditions and processing parameters. The toxicology library in UNIFI is comprised of over 1000 compounds including many PPCPs, such as drugs of abuse, veterinary medicines, and pharmaceuticals. Library entries also contain retention times and accurate theoretical fragment masses. Experimental conditions, sample preparation protocols, and data processing parameters are available in a previous application note by the same authors.2
From a previous application note,2 the screening of a local well water sample against the full toxicology library in UNIFI, with up to three adducts (H,+ Na,+ K+), indicated the presence of the four compounds in Table 1.
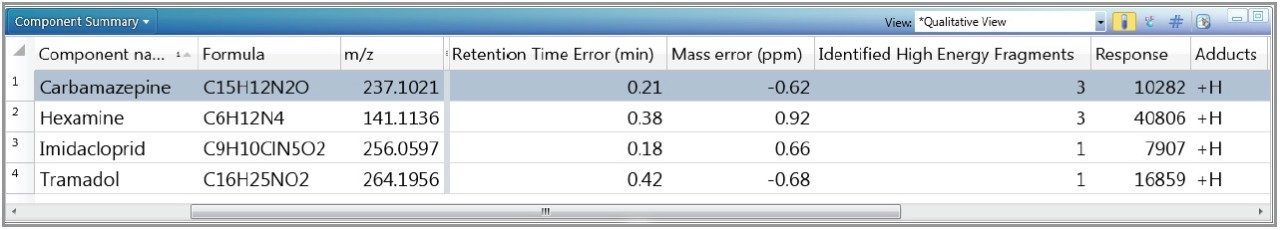
The inclusion of retention times and accurate mass fragment ions in the toxicology screening library allowed for confident matches to be made since they were based on more information other than accurate mass of the precursor ions alone. As indicated, this is critical for reducing false detection rates, enabling rapid data review for screening experiments.
Further investigation of the comprehensive dataset was possible using the metabolite identification functionality of UNIFI’s screening solution software. This functionality requires a target molecule with mol file and a list of possible transformations, that are shown in Figure 1.
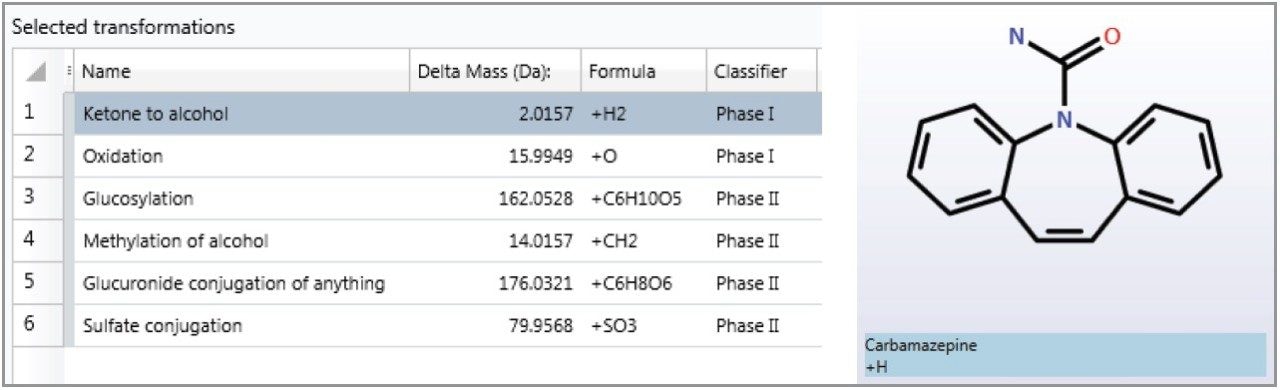
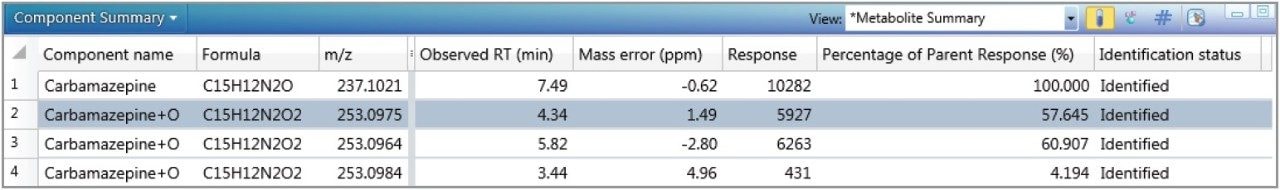
Primarily, using chemical intelligence,4 the target mol file is systematically cleaved. This essentially increases the target list to include parent compounds and potential breakdown products in the metabolite search.Interrogation of the low energy function of the MSE comprehensive dataset was performed, which automatically extracted the masses corresponding to the parent as well as the permutations of provided transformations, with and without systematic cleavages of the parent molecule. The list of possible metabolites for carbamazepine is shown in Table 2 and Figure 2.
No metabolites were observed for the other three compounds found in the screening experiment.
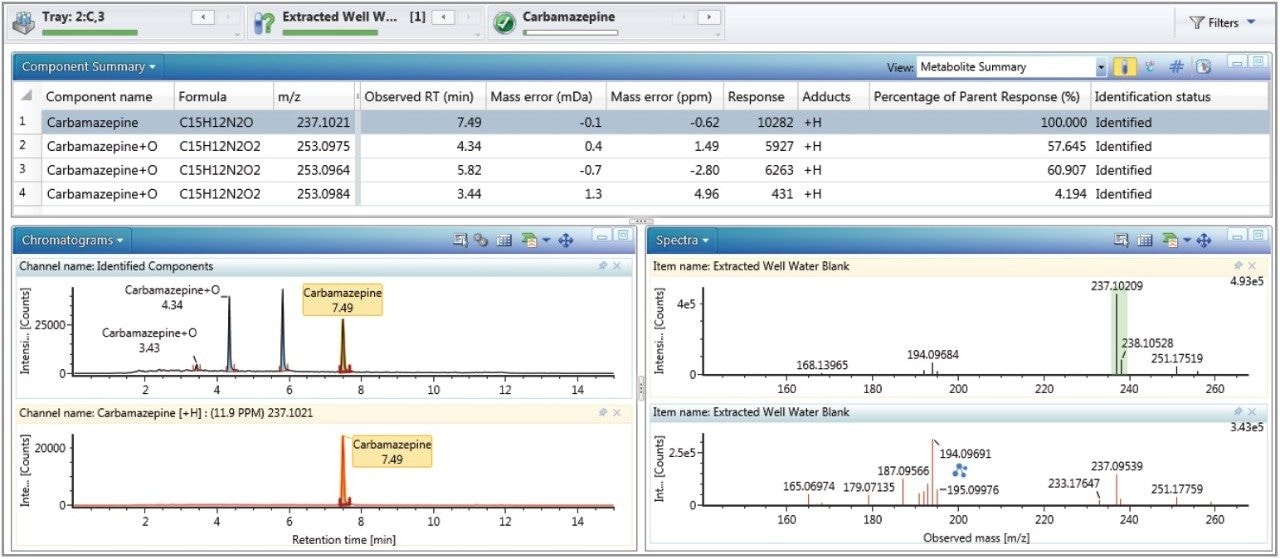
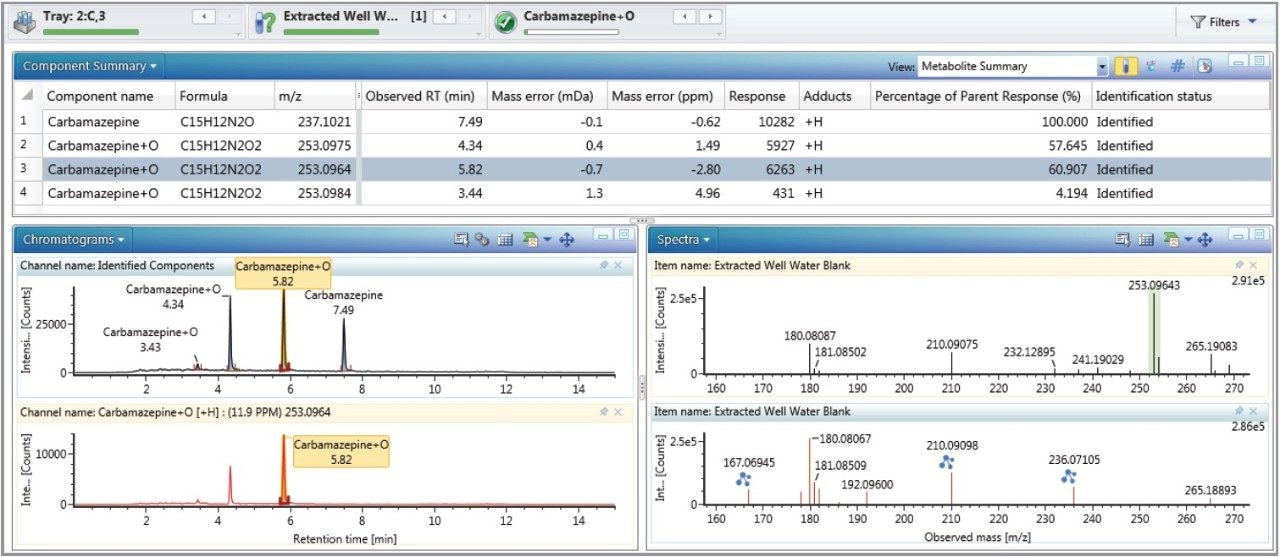
Figures 3 and 4 show the full UI information details for the identification of carbamazapine and a carbamazepine oxidation respectively. Fragment match functionality within UNIFI uses similar intelligence as the cleavage algorithm above. It systematically dissects the mol file of the parent or proposed metabolite and assigns potential accurate mass fragment ions from the high energy function of the MSE data. Identified fragment ions are annotated, as shown in Figure 3 for the mass 194.06691 Da, and in Figure 4 for the masses 210.09098 Da and 236.07105 Da.
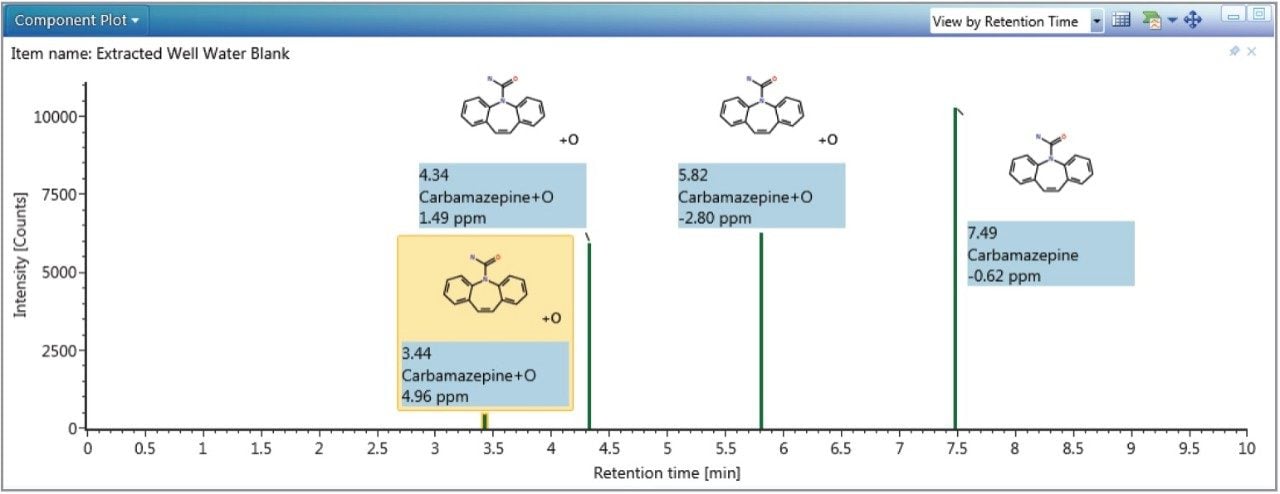
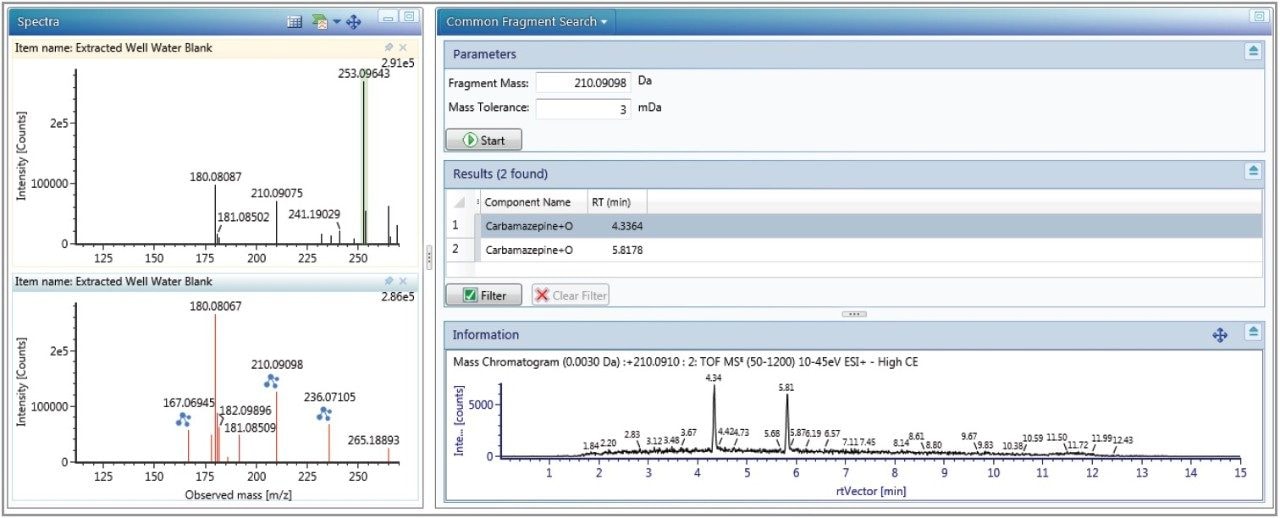
Just as in screening experiments, the high energy fragment ions provided increased confidence that identified metabolites were correct. Common fragment and neutral loss discovery tools, readily available in UNIFI, can also be used to enhance the confidence in metabolite identification. Figure 5 shows the results of running a common fragment search. The two +O metabolites of carbamazepine at 4.3 and 5.8 minutes are shown to be related to each other by the fragment 210.0910 Da, which is the loss of 43.005 Da from the parent 253.0964 Da. This is the same neutral loss from the carbamazepine parent (237.1021 Da) to the primary fragment (194.0969 Da) thus giving further confidence in the metabolites identified.
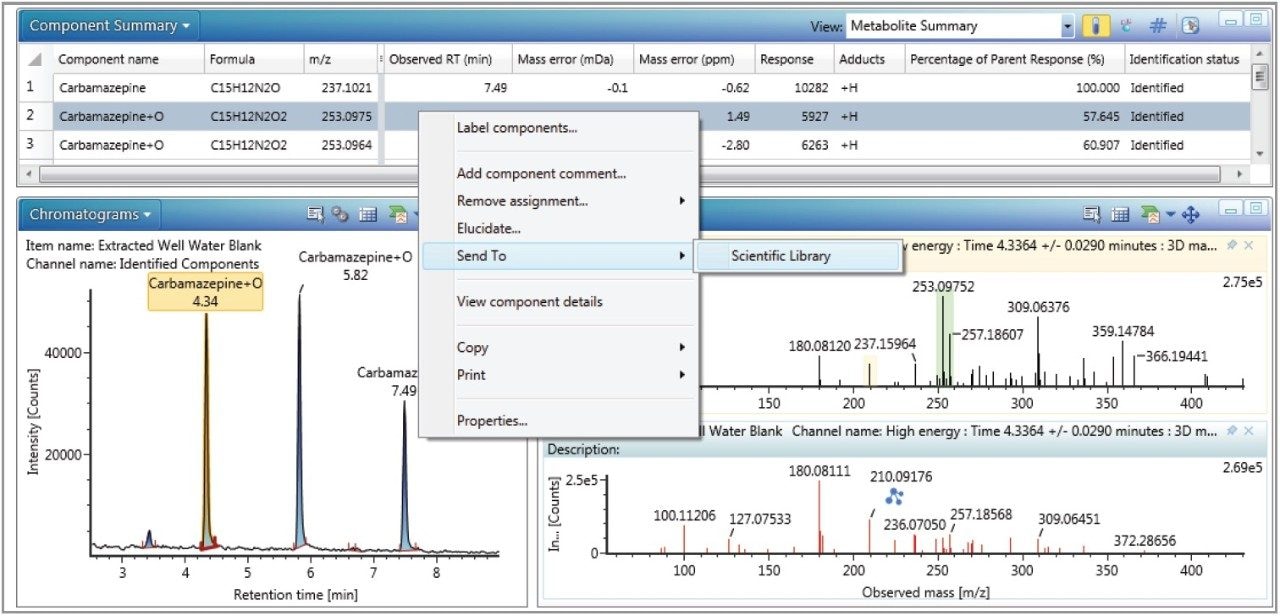
Once the presence of a metabolite has been confirmed, the entry can be easily exported to an existing or new scientific library within UNIFI with the right click of the mouse, as shown in Figure 6. Details such as formula, retention time, theoretical accurate mass fragment ions, and spectra are made available for future users and analyses.
720004812, July 2017