For research use only. Not for use in diagnostic procedures.
In this paper we have used two dimensional preparative electrophoresis to separate proteins in an extract from H. influenzae b.
Haemophilus influenzae is a major pathogen responsible for a wide variety of human infections. This virulent bacterium causes serious diseases such as meningitis, epiglottitis, bacteremia, cellulitis, acute otitis media, and sinusitis. The physiology of this pleomorphic, Gram-negative coccobacillus bacterium has been well characterized over recent years and it was the first genome of a free-living organism to be completely sequenced (Fleischmann et al., 1995).
The cell walls of H. influenzae contain lipooligosaccharide, which resembles the lipopolysaccharide of Gram-negative bacilli but has shorter side chains (hence the designation oligosaccharide rather than polysaccharide). H. influenzae does not make toxins or other extracellular products that account for their ability to produce infection. The mechanism of pathogenesis of H. influenzae infections is not completely understood. However, the presence of the type b polysaccharide capsule is a major factor in virulence.
Encapsulated organisms can penetrate the epithelium of the nasopharynx and invade blood capillaries directly.
The bacterium use pili and fibrils during the process of attachment and colonisation of the host. These cell surface macro structures contain proteins that may be potential vaccine candidates. However, identification of membrane associated proteins is difficult using traditional proteomics techniques such as 2D PAGE. This is because the membrane proteins contain hydrophobic transmembrane domains that span the phosopholipid membrane. The result of these hydrophobic domains is that the protein is difficult to solubilise and will not remain in solution during the isoelectric focusing on an IPG strip.
In this paper we have used two dimensional preparative electrophoresis to separate proteins in an extract from H. influenzae b.

Bacterial Cultivation and Protein Preparation
The H. influenzae b strain (D6) was cultivated in PDM antibiotic sensitivity medium (PDM, haemoglobin, isovitalex) at 37 °C in a low partial pressure of oxygen. The bacteria were harvested and the pellet was washed in PBS. Bacteria from a single culture dish were used.
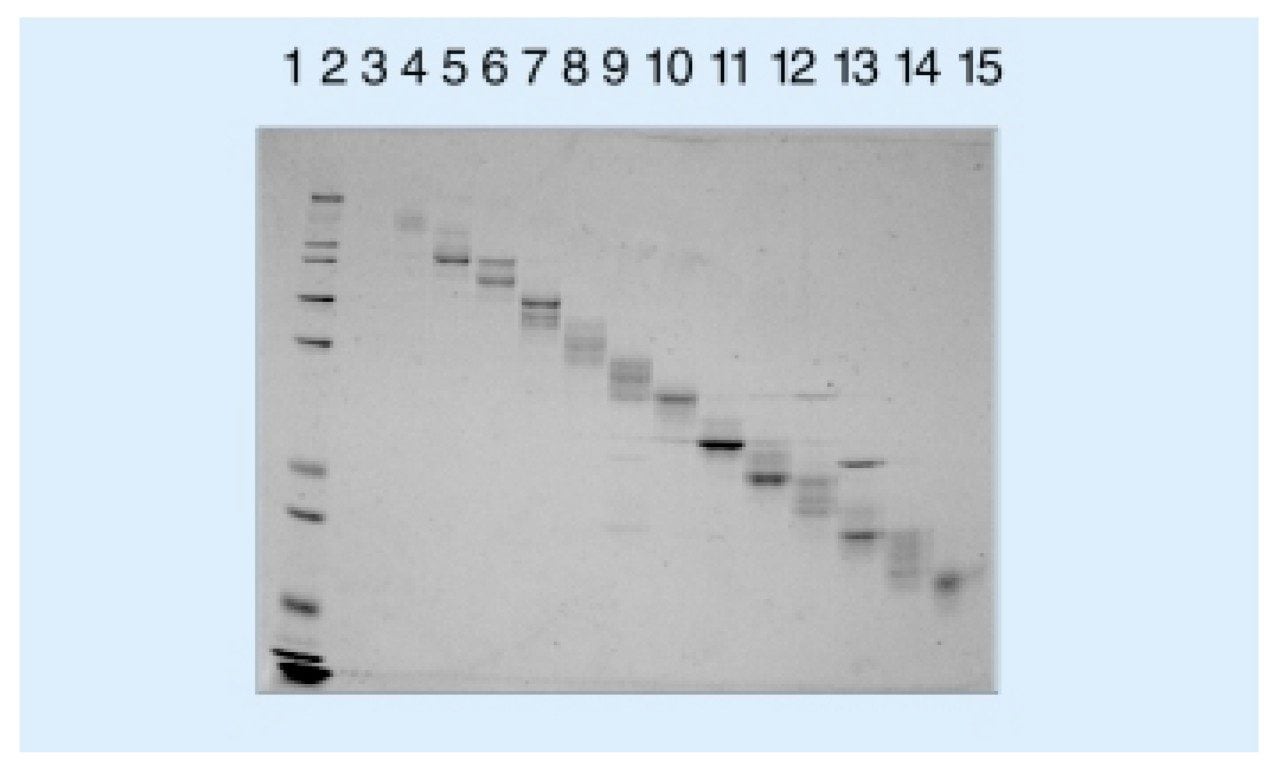
The samples were dissolved in 1 ml digitonin (0.1%) and ultra sonicated for 10 min. Another 14 ml of digitonin (0.1%) and servalyte (2%) was added to the sample. Liquid phase IEF was performed according to experimental procedures described in (1, 2). The sample was loaded into the (pre-cooled) Rotofor apparatus (Bio-Rad Laboratories, Hercules, CA) for fractionation in a wide range pH gradient (pH 3–10). Constant power (10W) was applied to the system. The initial voltage applied was 880V and the system was run until a voltage plateau was reached, after about 2 hours. Twenty fractions were and pH was measured. The fractions were analysed with the NuPAGE System which includes 10% Bis-Tris gels, 3-(N-morpholino)- propane sulfonic acid (MOPS) sodium dodecyl sulphate (SDS) running buffer system. The proteins were detected using colloidal Coomassie blue (Gel Code Blue, Pierce).
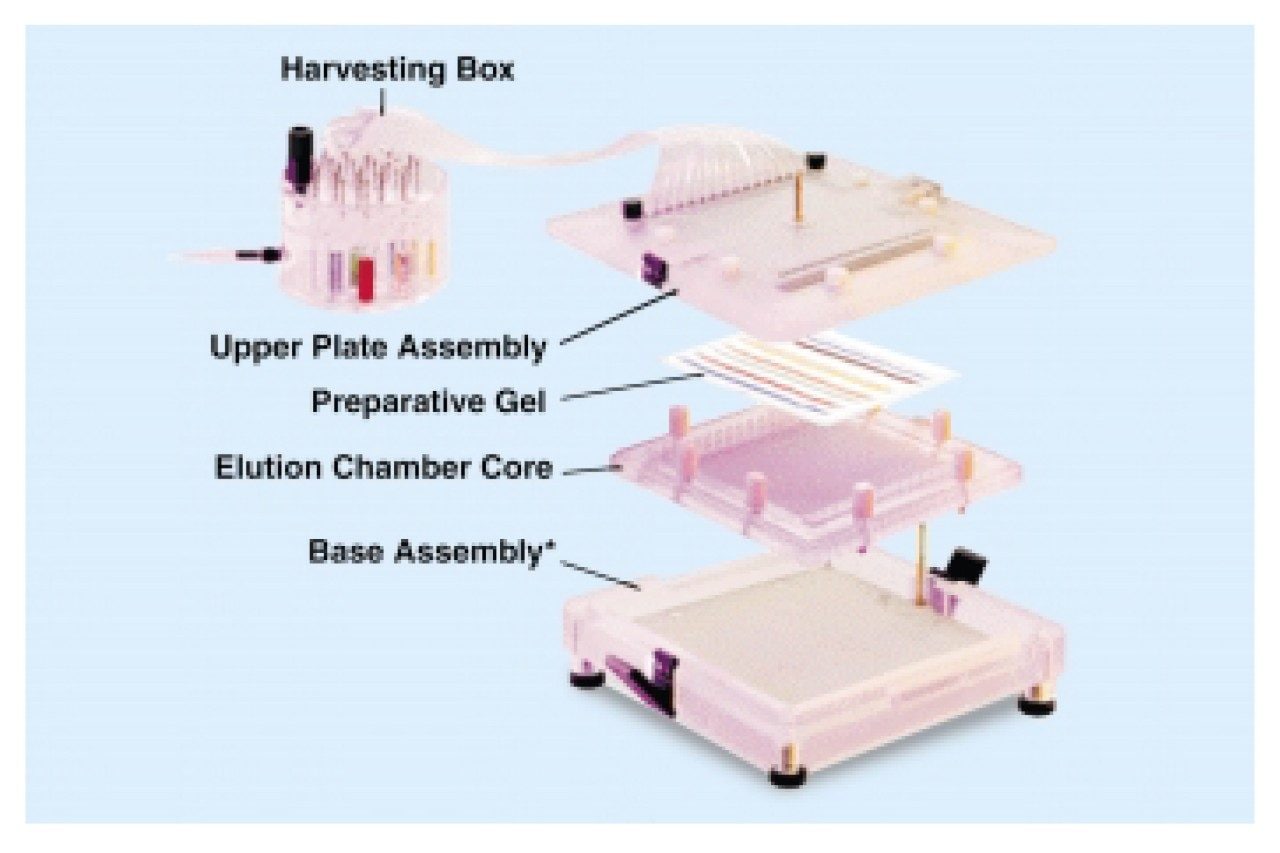
Rotofor fractions were concentrated by vacuum centrifugation, dissolved in 200 μL NuPAGE SDS sample buffer (Novex) and heated at 100 °C for 2–3 min. Samples were separated using the NuPAGE System run at 200V for 55 min. The fraction was then extracted using the mini whole gel eluter (Bio-Rad) following the manufacturer’s instructions from the whole gel eluter and mini whole gel eluter, (Davidsson, et al, 1999). The system was run at 50V.
Gel eluter fractions were dried to a volume of 200 μL. 600 μL of ice cold acetone was added and the samples were incubated in freezer for 2 hrs. The samples were centrifuged and dried after the supernatant was removed. The proteins were dissolved in digestion buffer (0.1 mM CaCl2, 0.1 M NH4HCO3) and 10 μL of trypsin (10 ng/μL) was added followed by incubation at 37 °C for 4 hrs. The samples were dried and dissolved in 25 μL TFA (0.1%).

MALDI-MS analysis was performed using a TofSpec 2E (Micromass). Dried tryptic digests were reconstituted in 25 μL 0.1% TFA and treated with C18 Zip Tips (Millipore) according to the manufacturer’s instruction. 0.5 μl sample was mixed with 0.5 μl matrix solution (α-cyano-4- hydroxy cinnamic acid 10 mg/mL in acetonitrile: H2O, 1:1) directly on the MALDI probe and allowed to dry under ambient conditions. Peptide spectra were acquired in reflectron mode at an accelerating voltage of 20 kV and 200 laser shots were summed. External calibration using angiotensin II and ACTH was used. MALDI-spectra were analysed using the MassLynx Software in a WindowsNT environment. Resulting values for monoisotopic peaks were searched against the NCBInr database.
Samples for which no confident identification could be made using peptide mass fingerprinting were analysed by nanoflow electrospray tandem mass spectrometry on a Q-Tof* (Micromass, UK). Samples were enriched using Zip Tips, eluted with acetonitrile:H2O containing 0.1 % formic acid, and sprayed from coated borosilicate capillaries (Micromass). Doubly and triply protonated peptides were fragmented using argon as a collision gas. Instrument calibration was performed using fragment ions from Glu-fibrinopeptide B and a fourth-order polynomial fit. Fragment ion spectra were post-processed in a standard text file format and searched in the NCBI nr database.
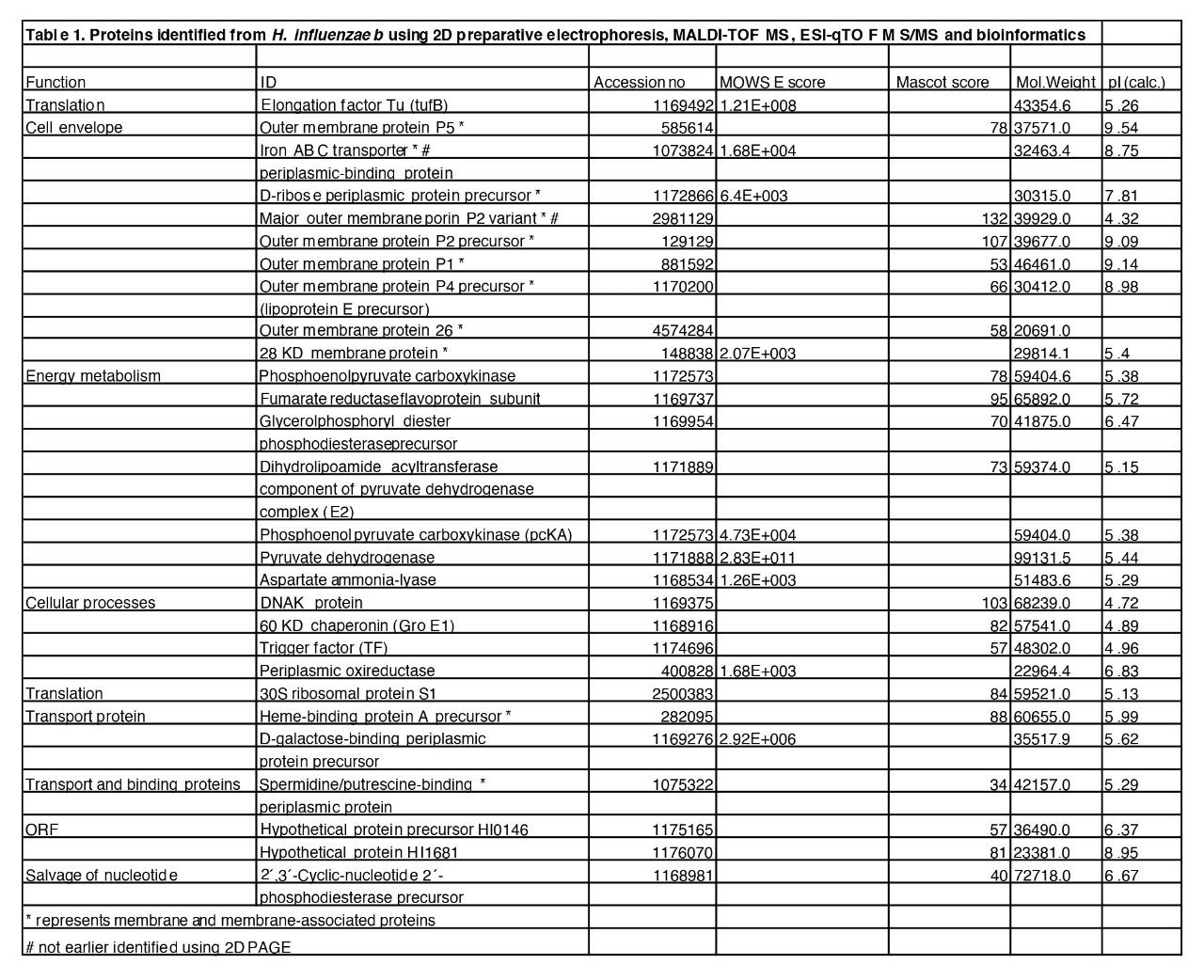
We have used a rapid 2D preparative electrophoresis technique to isolate 30 proteins from H. influenzae b and identify them using MALDI-TOF MS, ESI-QTOF MS/MS, and bioinformatics. Out of the 28 proteins 11 (39%) were identified as membrane or membrane- associated proteins. To the best of our knowledge two proteins not previously identified using 2D PAGE were identified using preparative electrophoresis as a separation technique. This method, which is rapid and requires no prefractionation, can be used as a complement to 2D PAGE in studies of protein expression.
* Clayton E., Bateman R.H. Time-of -flight Mass Analysis of Highenergy Collision induced Dissociation Fragment Ions. Rapid Communications in Mass Spectrometry 1992, 6:719.
Morris H.R., Paxton T., Dell A., Langhorne J., Berg M., Bordoli R.S., Hoyes J., Bateman R.H. High Sensitivity Collisionally Activated Decomposition Tandem Mass Spectrometry on a Novel Quadrupole/Orthogonal-Accelleration Time-of-Flight Mass Spectrometer. Rapid Communications in Mass Spectrometry 1996, 10:889–896.
720000555, February 2001