This is an Application Brief and does not contain a detailed Experimental section.
The use of BioAccord LC-MS System combined with a novel informatics workflow for peptide MAM analysis.
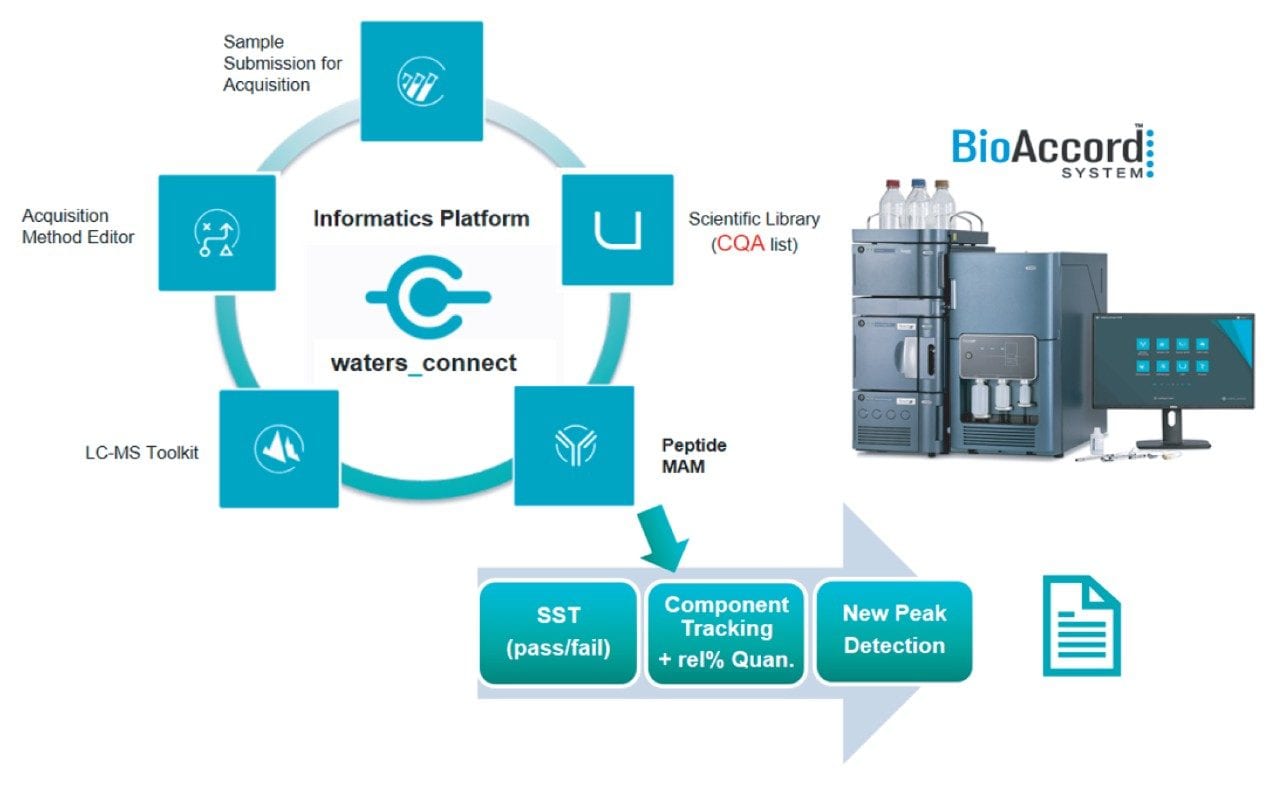
In recent years there has been a concerted effort by the pharmaceutical industry to deploy multi-attribute method (MAM) for different stages of the product life cycle, such as clinical material characterization, stability testing, and QC product release. However, the successful deployment of the MAM is limited by challenges such as ease-of-use at the system level, method robustness, and reliability of systems deployed for this assay. Workflow steps such as peptide attribute tracking, relative quantitation, and new peak detection (NPD) are also expected to meet strict compliance and usability requirements. In this technology brief we introduce a new MAM workflow that extends the capabilities of the BioAccord LC-MS System with a dedicated peptide MAM application operating under the waters_connect compliant-ready informatics platform. This new analytical workflow enables organizations to obtain reliable relative quantitation of pCQAs (product critical quality attributes) and detection of potential unknown impurities at peptide level. The integrated end-to-end workflow design improves the overall user experience with streamlined instrument operation and automated MAM data processing while providing integrated data acquisition, processing, and reporting within regulated and non-regulated laboratory environments.
The BioAccord LC-MS System, the first with SmartMS Technology, operates under the compliant-ready waters_connect Informatics platform (Figure 1). The waters_connect platform supports multiple integrated application workflows for biotherapeutic analyses from intact mass analysis to released glycan profiling and peptide attribute characterization and monitoring. Figure 1 illustrates workflow steps for peptide MAM analysis from LC-MS method development to execution of an assay for peptide attribute monitoring and quantitation. The peptide MAM application has been designed to carefully address the challenges of data processing from managing system suitability test (SST) injections to the processing of blanks, control/reference, and analytical samples. The SST ensures the readiness of the LC-MS system for the analytical workflow to be performed. The application further tracks peptide attributes of interest and reports relative %modification levels of targeted pCQAs for each sample. User definable thresholds can be applied to both system suitability tests and peptide monitoring data to clearly flag attributes and analyses exceeding those predefined limits. During purity assessment, a new peak detection algorithm identifies novel impurities and ions changing substantially in analytical samples compared to the reference.
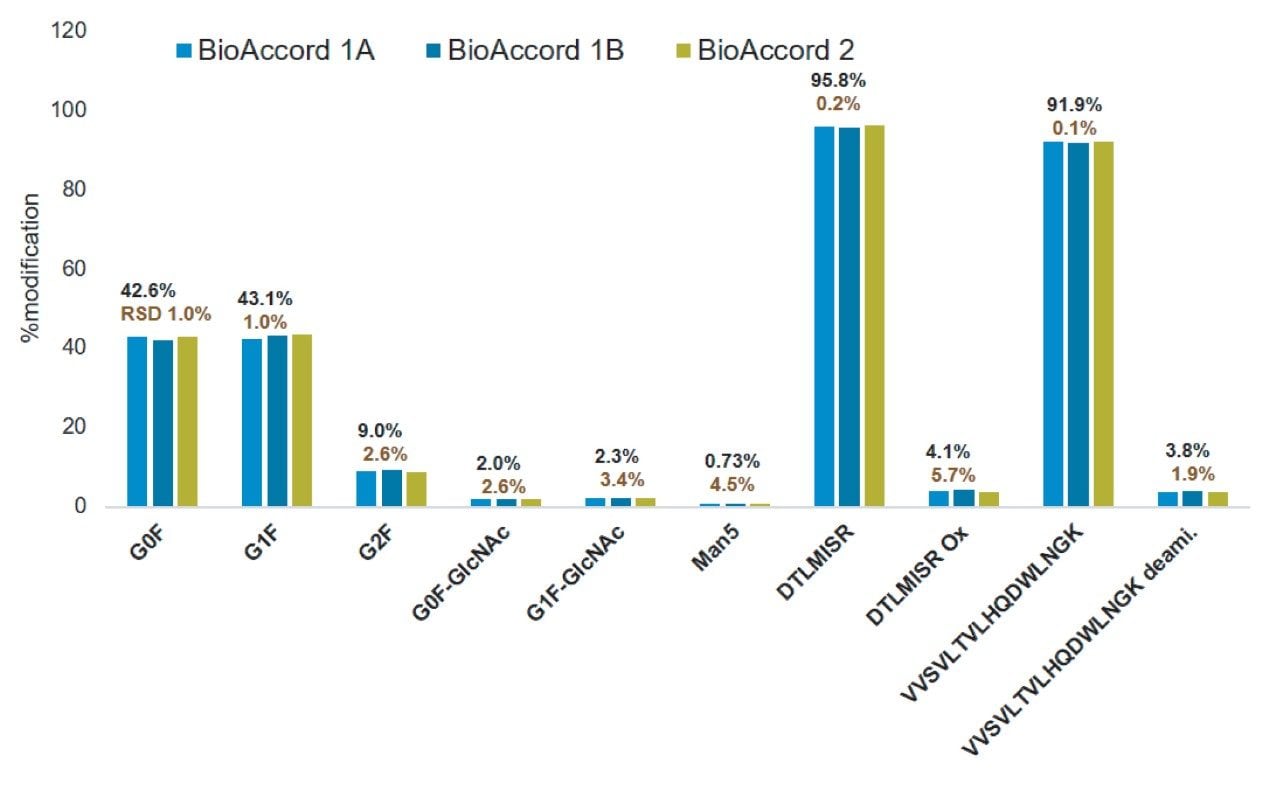
The deployability of MAM across an organization depends on the ability to generate reproducible data not only within a system but across systems with different operators. To measure the performance of the BioAccord System and waters_connect platform together for execution of the peptide MAM workflow, an analysis was performed for a set of system suitability peptides: 1a) control sample which is a tryptic digest of the NISTmAb reference, 1b) control sample spiked in with a quantitative peptide mixture, 2a) stressed sample: NISTmAb reference standard that was heat and pH stressed and tryptic digested, 2b) stressed sample spiked in with a quantitative peptide mixture. On two separate BioAccord systems the MAM analysis was repeated from two weeks to three months apart. Figure 2 shows the results from the two systems, yielding inter- and intra-system reproducibility with <6% relative standard deviation (%RSD) for monitored %pCQAs, suggesting that the MAM study can be reproduced in its entirety across multiple systems.
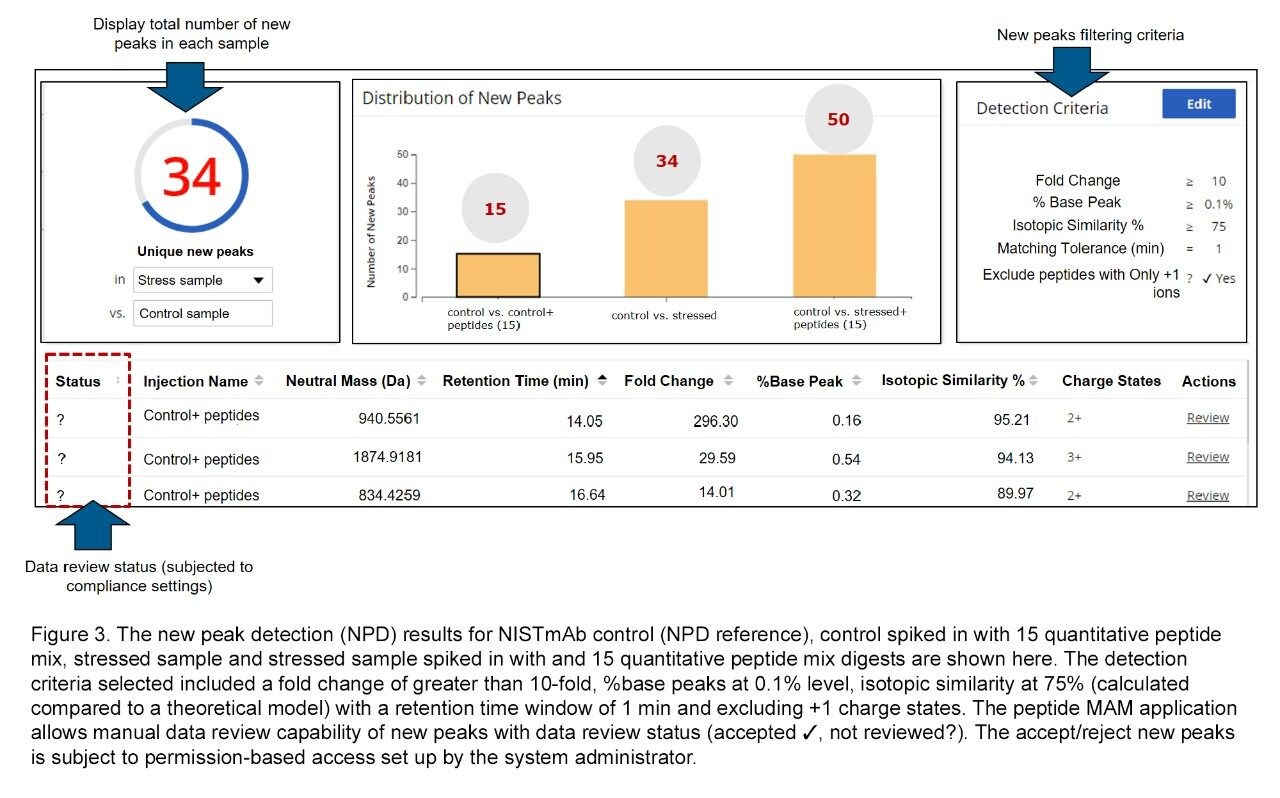
New peak detection (NPD) is an essential part of peptide MAM analysis when choosing to use the method as a purity assay for a biotherapeutic. The NPD processing flags novel impurities in the analytical samples that are or those that have changed substantially compared to a reference. The criteria for new peak detection often change from laboratory-to-laboratory and molecule-to-molecule, therefore the data processing for NPD requires flexibility in user defined settings to establish thresholding parameters. The parameters can be further modified following data processing to filter new peaks from potential false positives. The newly introduced %isotopic match criteria filter significantly reduces detection of these false positives and increases the confidence of flagged peaks as potential impurities (Figure 3).
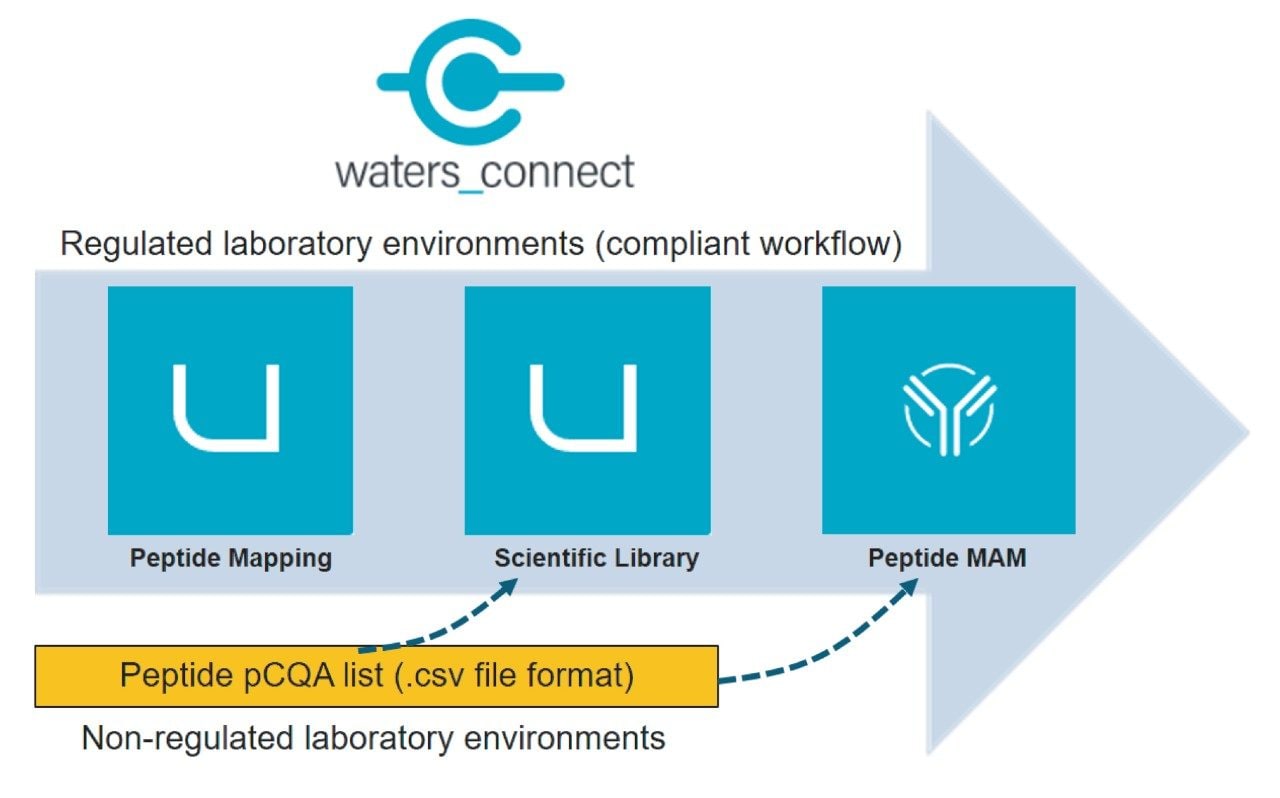
The pCQAs for attribute tracking and monitoring that are defined during peptide mapping characterization studies can be introduced to the peptide MAM processing method through “add” or “import” functions. For assembling the monitoring list of peptides, the “import” function provides the most streamlined process as pCQAs can be directly imported into the MAM method from a waters_connect scientific library populated from the characterization run. This integrated process also addresses data traceability and potential compliance expectations from regulated organizations. However, to support attribute list imports from multiple informatics platforms or vendor systems, the CQA library can also be populated in the scientific library using a .csv import file functionality (Figure 4).
Here we have presented the key elements of the new peptide MAM workflow that extend the capabilities of the BioAccord LC-MS System coupled with the novel waters_connect informatics platform. This innovative platform design with multiple integrated applications for attribute characterization and monitoring complements the usability of the SmartMS-enabled BioAccord instrument system. The workflow further provides quick turnaround time to results enabling wider access to high information content for faster and better decision making.
720006963, July 2020