For research use only. Not for use in diagnostic procedures.
A HILIC-UPLC separation with ion mobility-Tof MS (SYNAPT G2-S HDMS) enables a multi-dimensional separation of complex biological mixtures, enhancing the information obtained from profiling lipids. HDMS Compare Software and TransOmics Informatics facilitate the comparison of the biological samples.
The combination of liquid chromatography, ion mobility, and oa-Tof mass spectrometry is a multi-dimensional separation strategy capable of analyzing complex biological mixtures to a depth not previously possible, enhancing the detail obtained from lipidomic profiling.
Combining HILIC-UPLC liquid phase separation with gas phase ion mobility mass spectrometry to achieve a multi-dimensional characterization of lipids in complex mixtures enhances profiling of lipids in biological samples.
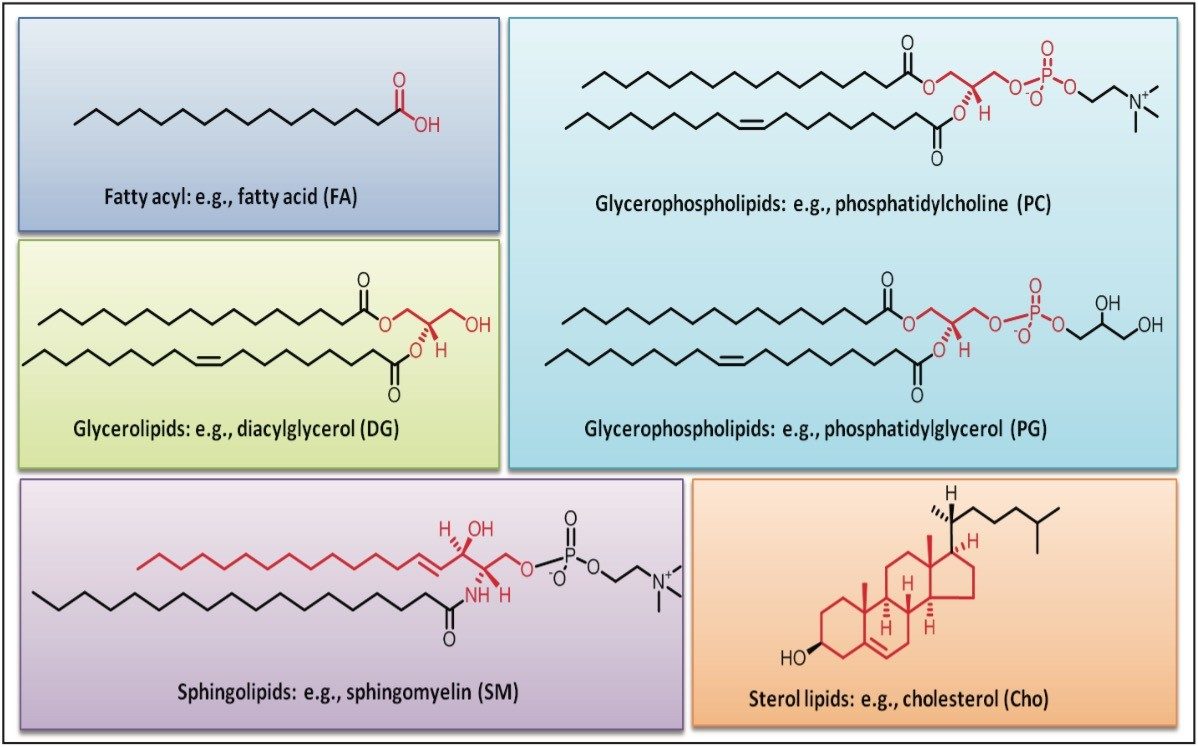
One of the main challenges for a global lipid analysis (lipidomics) is the separation of the wide array of lipid species present in biological samples (Figure 1). Such a separation is not achievable using a single chromatographic dimension such as reversed- or normal-phase separation methods.1-5 Normal-phase UPLC separates lipid classes based on their polar head group, whereas reversed-phase separates lipids according to their acyl chain length and number of double bonds.1-6 Hydrophilic interaction chromatography (HILIC) separation has been proposed as an alternative to normal-phase separation, offering better MS compatibility and using less toxic solvents.4-6 Recently, a two-dimensional separation using HILIC and reversed-phase has been proposed to maximize the separation of the lipidome before MS detection.5,6
In addition to chromatography, ion mobility can be used to separate lipid ions in the gas phase according to their size and molecular shape.7,8 In this study, we apply the Waters Omics Research Platform with TransOmics Informatics. A HILIC-UPLC separation with ion mobility-Tof MS (SYNAPT G2-S HDMS) enables a multi-dimensional separation of complex biological mixtures, enhancing the information obtained from profiling lipids. HDMS Compare Software and TransOmics Informatics facilitate the comparison of the biological samples.
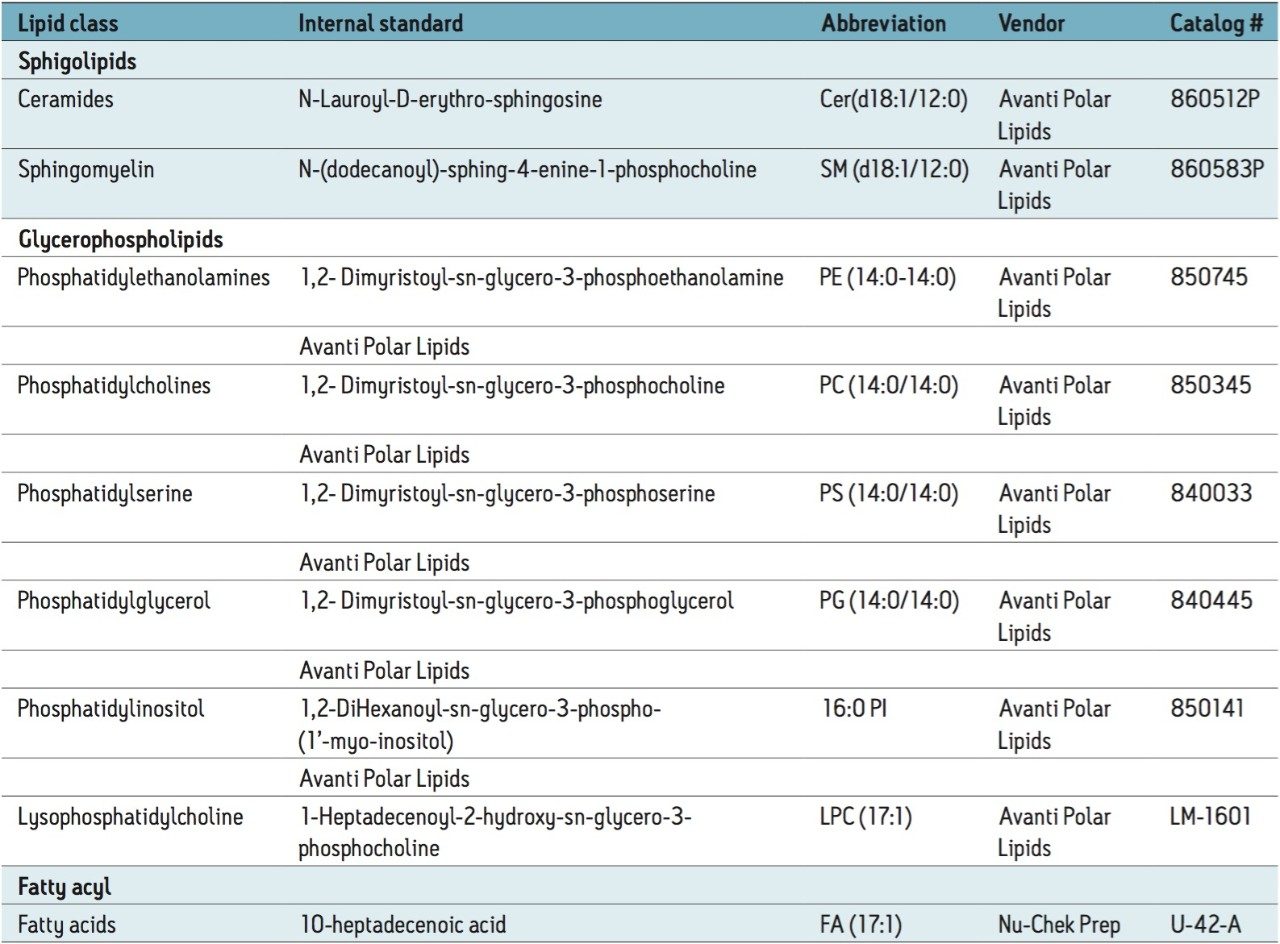
Lipid standards and total lipid extracts from bovine brain, heart, and liver were purchased from Avanti Polar Lipids. Non-natural lipids were spiked in the biological extracts and used as internal standards (Table 2).
|
System: |
ACQUITY UPLC |
|
Column: |
ACQUITY UPLC BEH HILIC 2.1 x 100 mm |
|
Column temp.: |
30 °C |
|
Mobile phase A: |
10 mM ammonium acetate (pH 8.0) in 95% ACN |
|
Mobile phase B: |
10 mM ammonium acetate (pH 8.0) in 50% ACN |
|
Time/min |
%A |
%B |
|---|---|---|
|
0.00 |
99.9 |
0.1 |
|
10.00 |
80.0 |
20.0 |
|
13.00 |
20.0 |
80.0 |
|
13.01 |
99.9 |
0.1 |
|
16.00 |
99.9 |
0.1 |
|
Flow rate: |
0.5 mL/min |
|
|
Injection volume: |
5 μL |
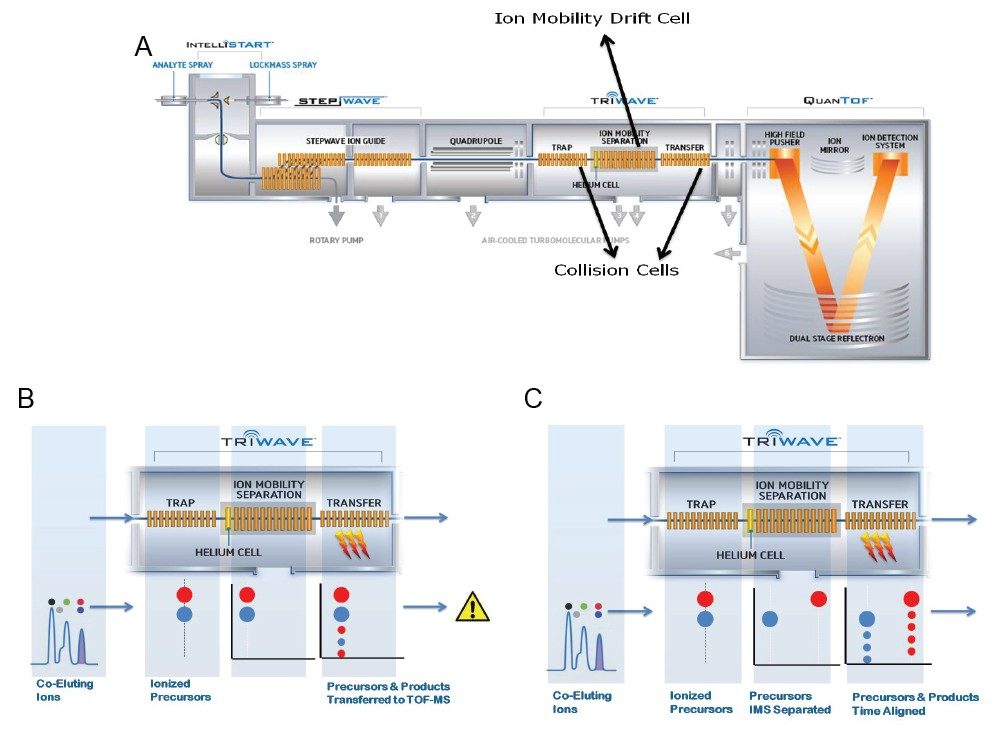
MS analyses were performed on a SYNAPT G2-S HDMS (Figure 2) with a conventional ESI source in LC-HDMSE mode. Capillary voltages were optimized separately for positive (2.8 kV) and negative (1.9 kV) ion modes. Data were collected in two channels all of the time; low collision energy (6.0 V) for the molecular ions and high collision energy (20 to 35 V) for product ions. IMS gas: nitrogen; IMS T-Wave velocity: 900 m/s; IMS T-Wave height: 40 V.
TransOmics Informatics and HDMS Compare Software
To separate lipids, we used hydrophilic interaction chromatography (HILIC) with an ACQUITY UPLC BEH HILIC 2.1 x 100 mm, 1.7 μm Column, and a reversed-phase solvent system (organic/aqueous) characterized by high organic mobile phase (>80% acetonitrile). This UPLC method was highly compatible with ESI, and separated lipids by classes, according to their polar properties (Figure 3 and Table 1).
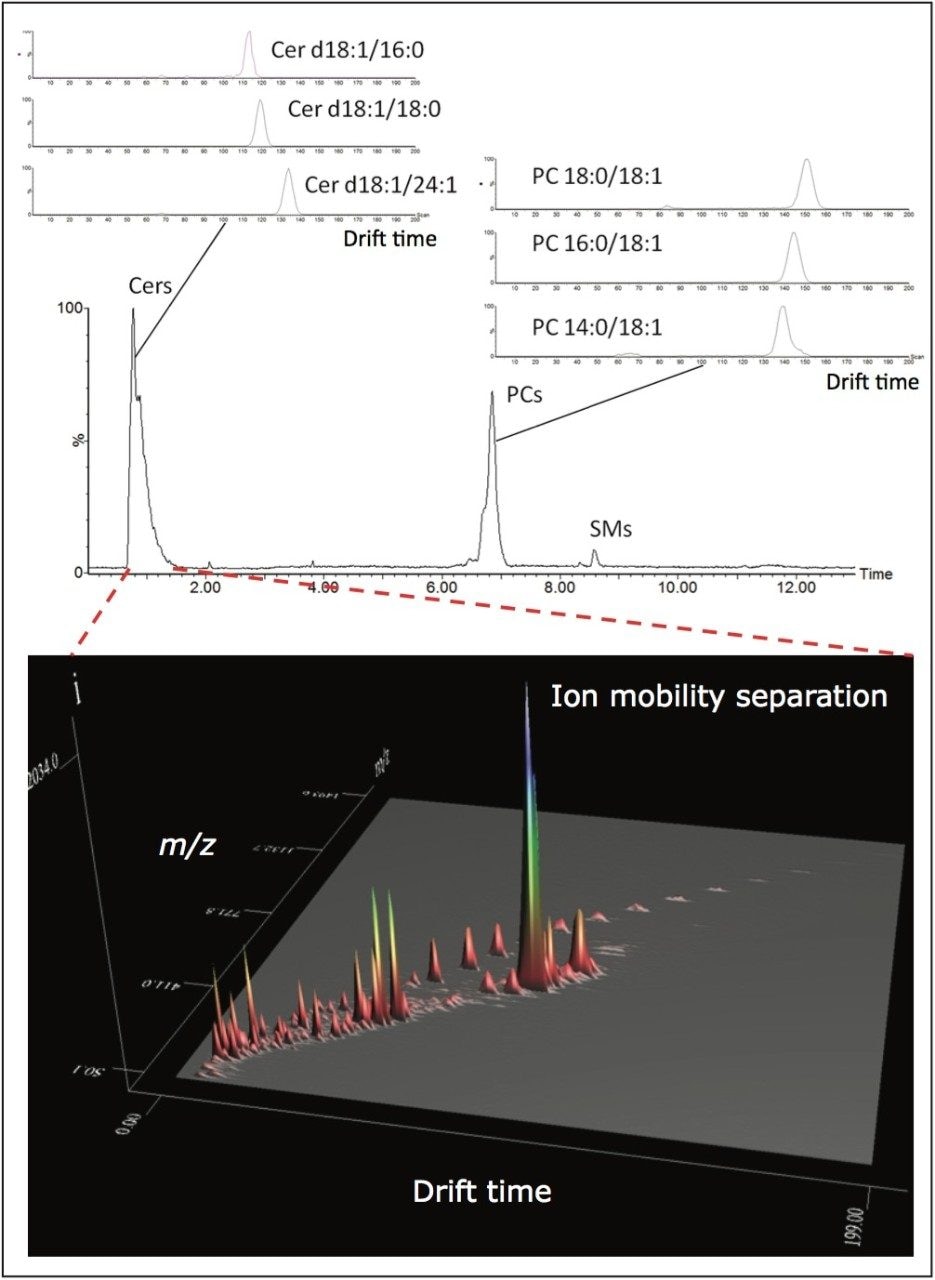
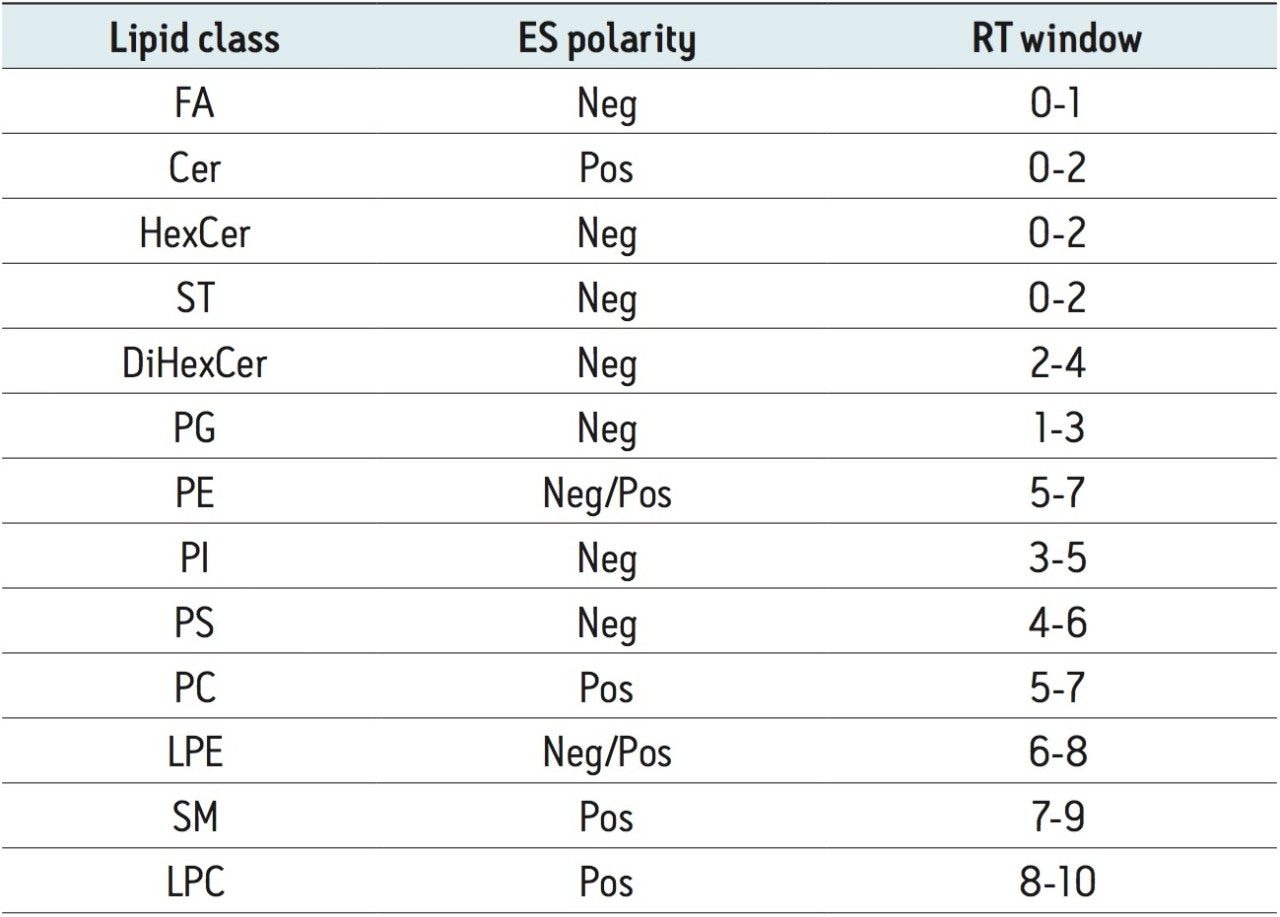
Table 1. Lipid classes are separated by retention time (RT) windows in HILIC conditions.
Abbreviations: FA, fatty acids; Cer, ceramides;
HeXCer, HexosylCeramides; ST, sulfatides;
DiHexCer, DihexosylCeramides;
PG, phosphatidylglycerols;
PE, phosphatidylethanolamines;
PI, phosphatidylinositols;
PS, phosphatidylserines; PC, phosphatidylcholines;
LPE, lysophosphatidylethanolamines;
SM, sphingomyelins; LPC, lysophosphatidylcholines.
In addition to HILIC chromatography, the ion mobility capability of the SYNAPT G2-S HDMS Mass Spectrometer (Figures 2A-C and 3) was used to further discriminate lipid classes into their constituent components, based upon the different size and shape, that is, the ions collision cross section (Ω).7,8 Lipid ions with different degrees of unsaturation and acyl length migrate with characteristic mobility times, due to their unique shape in the gas phase as they migrate through the ion mobility cell, which is filled with nitrogen gas at relatively high pressure (Figures 3 and 5). Ion mobility separations occur in the millisecond timeframe, making it ideal for situating between LC and MS, where LC separations upstream typically work in the second timeframe and Tof MS downstream works in the nanosecond timeframe (Figures 2A and 3). The addition of ion mobility to the LC-MS provides enhanced peak capacity and improved signal-to-noise ratio (Figure 3).
To gain more structural information, we analyzed lipids employing LC-MSE, which uses an alternating low and elevated collision energy in separate scans to acquire both precursor and product ion information in a single analytical run (Figures 2B and 4A). Ion mobility separation coupled with LC-MSE (HDMSE) improves the specificity for coeluting lipids by fragmenting ions after IMS separation (Figures 2C and 4B). Due to the complexity of the lipidome, the addition of ion mobility drift time as an orthogonal measurement to retention times provides complementary information regarding the lipid species, adding further specificity to lipid identification and data interpretation (Figure 4B).
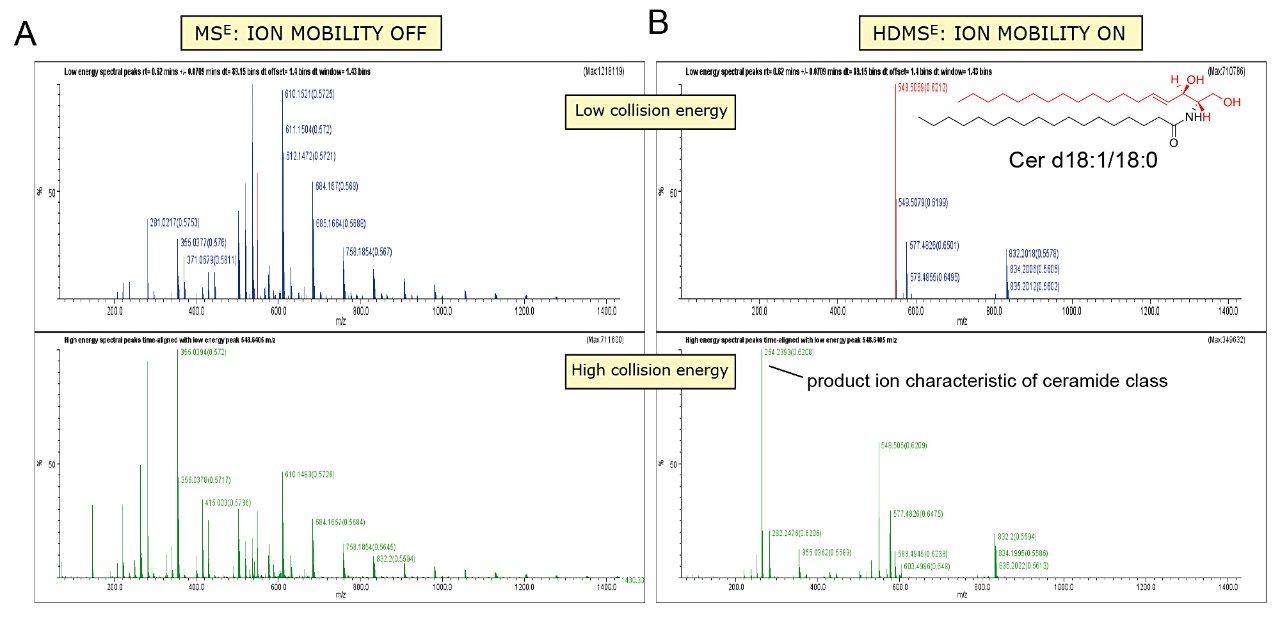
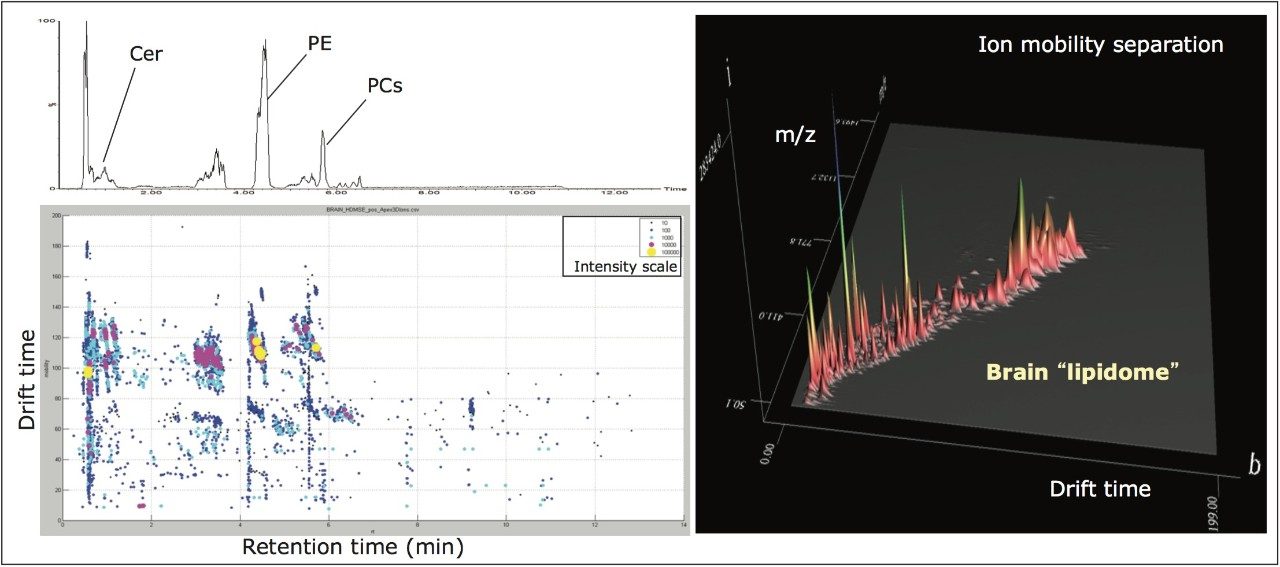
Using this novel technological approach, multidimensional molecular maps of lipids present in various animal tissues were generated. In these maps, each lipid is characterized by a combination of molecular coordinates including retention time, drift time, exact mass, fragment ions, and intensity (Figure 5). Such features highlighted the capacity of ion mobility to separate isobaric lipid species (i.e., species with the same mass). The molecular landscape visualized using multidimensional molecular maps also allows the detection of lipid species that could otherwise go unnoticed (Figures 3 and 5).
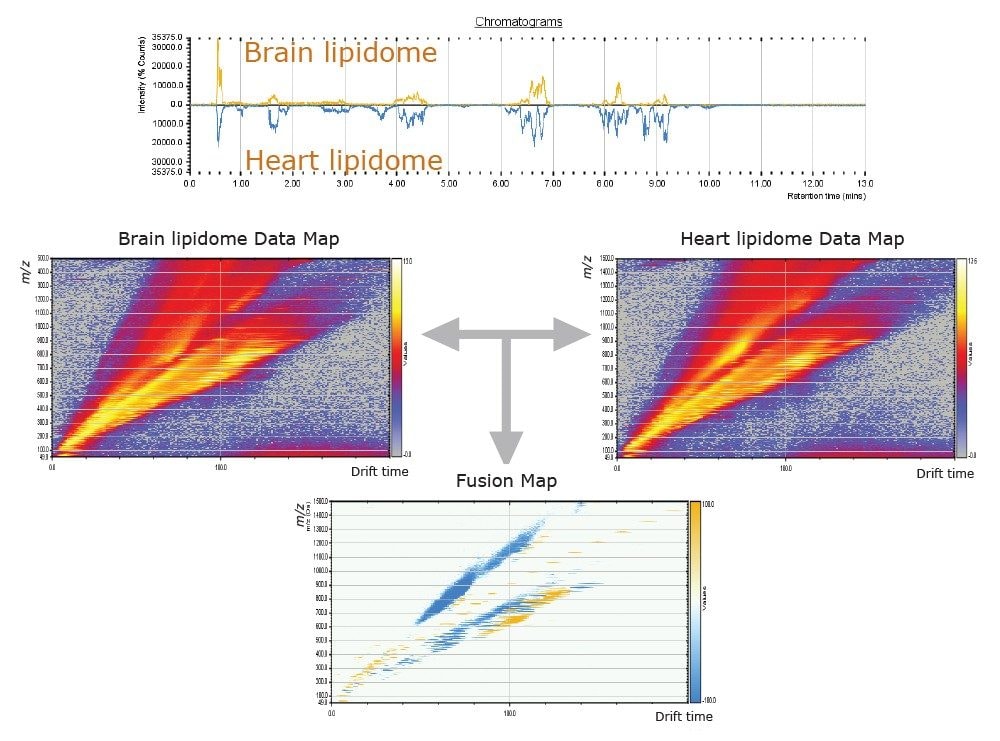
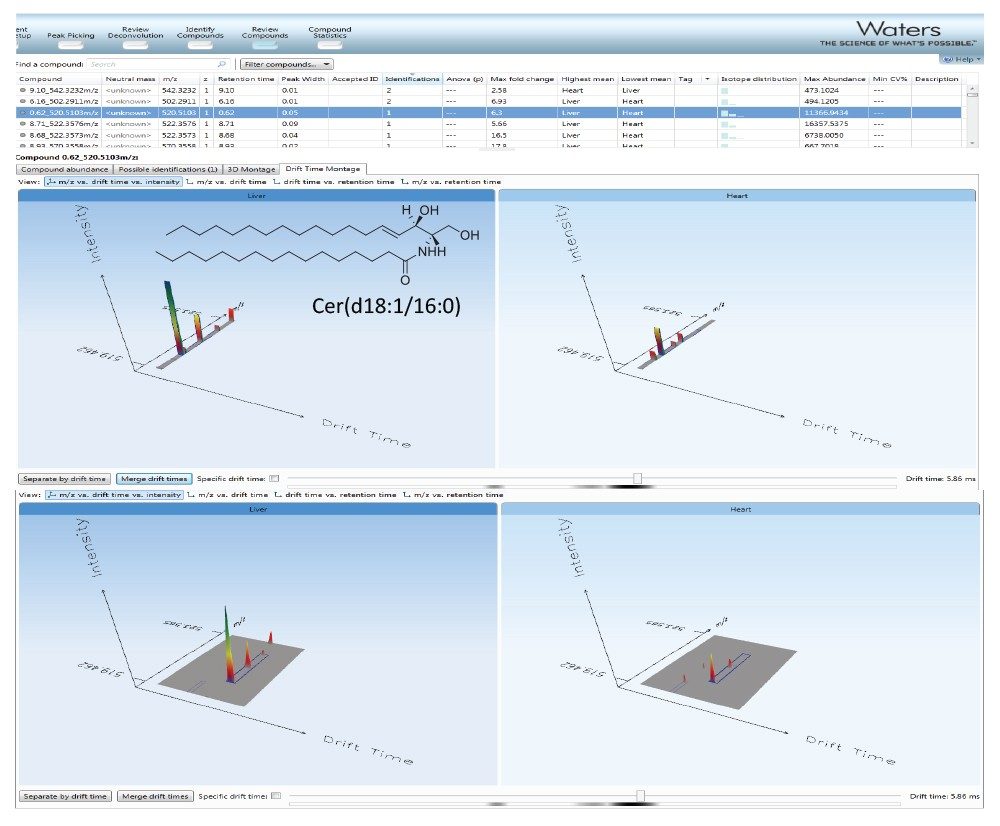
The comparison of molecular maps is facilitated by the use of HDMS Compare and TransOmics (Figures 6 and 7). HDMS Compare Software was used for a rapid comparison of different drift versus m/z plots at selected windows of retention times (Table 1). The drift time and spectral information associated with the components responsible for the differentiation can be extracted from the dataset and further analyzed (Figure 6). The use of TransOmics Informatics allows feature detection, alignment, and comparions across multiple samples using multivariate statistical approaches (PCA, dendrogram analysis) and database searching of discriminating features for the identification of the lipids alternating between samples. TransOmics uses ion mobility information to separate co-eluting isobaric lipids in the drift time dimension, increasing the specificity of identification and quantification (Figure 7). Lipid quantification was performed using appropriate internal standards for each lipid class (Table 2).
The combination of liquid chromatography, ion mobility, and oa-Tof mass spectrometry is a multidimensional separation strategy capable of analyzing complex biological mixtures to a depth not previously possible, enhancing the detail obtained from lipidomic profiling.
720004704, May 2013