Platform Ion Pairing RPLC Method for Oligonucleotides Using High Throughput 20 mm Length Columns
Abstract
A novel generic (platform) method is proposed for the separation of oligonucleotide size variants (10–100-mer) using ultra-short ACQUITY™ Premier Oligonucleotide BEH™ C18 2.1 x 20 mm columns. The proposed non-linear (logarithmic-like) mobile phase gradient overcomes the inherent problem of separating homologous compounds (e.g.: oligonucleotides and their short mer impurities) and affords the most uniform peak pattern distribution possible. With the initial and final mobile phase compositions determined, a generic 3–5 segment multi-linear gradient (approximating a logarithmic curve) is programmed that enables ultra-fast (1–3 minute) oligonucleotide separations on a routine basis.
Benefits
- Ultra-short (2.1 x 20 mm) columns allow for development of fast one to three minute oligonucleotide separations that maintain high efficiency and resolution
- Selectivity can be adjusted and maximized by programing non-linear (logarithmic-like) mobile phase gradients
- The logarithmic-like gradient establishes the basis for a universal (platform) method for separating oligonucleotides in the 10 to 100-mer size range
Introduction
Ion-pair reversed-phase liquid chromatography (IP-RPLC) performed at elevated temperature in the presence of an organic co-solvent is considered the method of choice for the analytical-scale separation of oligonucleotides (ONs).1 In principle, RPLC is suitable for liquid chromatography-mass spectrometry (LC-MS) analysis of oligonucleotides as well as nucleic acids which is a huge benefit compared to other chromatographic modes.1 RPLC provides the highest separation efficiency (peak capacity) for oligonucleotides when compared to any other chromatographic technique. In current practice, IP mobile phases consist of volatile alkylamines buffered with volatile acids (acetic acid or hexafluoroisopropanol (HFIP)). Although the adsorption and elution mechanism are quite complex (combined hydrophobic and electrostatic interactions operating via a multi-layer adsorption process), the measurements are reproducible and robust. Moreover, simple semi-empirical retention models can be used to design the separation and predict retention and selectivity.
Routine methods typically use 50 to 150 mm columns and typical run times are between 15 and 40 minutes.2,3 As shown in recent publications, ONs follow a similar elution behavior as other large analytes (peptides, proteins, antibody-drug-conjugates…), and their retention is very sensitive to the mobile phase composition.4 Oligonucleotides essentially get trapped at the head of the column until a small change in mobile phase strength triggers their complete elution without any further interaction. Column length has therefore little impact in controlling their retention, allowing for shorter columns to be considered. Known as the “bind-and-elute” or “on-off” elution mechanism, it contrasts with the multistep partitioning process that governs the elution of small molecules.
In fact, in most cases, the on-off elution behavior is explained by the large size (mass) of the solute. Larger polar molecules are generally more "rejected" by a less polar solvent (i.e. RPLC eluent including acetonitrile or methanol) - this assumption is based on Horváth's solvophobic theory.5 In addition, Regnier describes the on-off mechanism in terms of the number of displacing solvent-eluent molecules (stochiometric displacement model).6 Other theories explain this particular elution behavior with the solvation of the analyte and the size of the solvation layer surrounding the analyte. For oligonucleotides, despite their relatively small size, the likely reason for their on-off elution mechanism is their chain-like structure. ONs are linear molecules and therefore their binding surface is relatively large compared to their size/mass. On the other hand, since they are very polar, it is reasonable to assume that they are surrounded by a thick layer of water molecules. In addition, ion pairs are formed which also increase their apparent size.
In this application note, we propose a generic approach to develop platform methods for the fast analysis of ONs with length variants in the size range of 10 to 100-mer. Our approach is based on the “homologue rule” - like retention pattern of ONs and the so-called “inverse gradient function” methodology.7,8
Experimental
LC Analysis of Oligonucleotide Ladders
Sample preparation:
An oligo dT ladder was prepared by reconstituting the contents of a vial of MassPREP™ Oligonucleotide Standard (p/n: 186004135) in 100 µL water. Two ssDNA ladders were prepared by reconstituting the contents of a vial of ssDNA Ladder 10–60 Ladder (p/n: 186009449) and a vial of ssDNA Ladder 20–100 Ladder (p/n: 186009448) in 100 µL water.
Mobile phase preparation:
HPLC grade water and acetonitrile (MeCN) were obtained from Fisher Scientific (Dublin, Ireland). Triethylamine (TEA), diisopropylethylamine (DIPEA), hexylamine (HA), octylamine (OA), and hexafluoroisopropanol (HFIP) were purchased from Sigma- Aldrich (Buchs, Switzerland).
HFIP buffer was prepared as 100 mM solution, then 10 mM DIPEA was added, and pH of the solution was adjusted to ~8.4. This solution was used as mobile phase A. For mobile phase B, the 100 mM HFIP plus 10 mM DIPEA was prepared in a mixture of MeCN and water 1/1 (v/v).
|
LC system: |
ACQUITY UPLC™ H-Class Bio System with Binary Solvent Manager [Equivalent to an ACQUITY Premier System with BSM FTN instrument configured with a High pH Kit] |
|
Detection: |
ACQUITY TUV Detector (Titanium Flow Cell, 5 mm, 1500 nL) |
|
Wavelength: |
260 nm |
|
Data acquisition: |
Empower™ Pro 3 Software Feature Release 3 |
|
Column: |
ACQUITY Premier Oligonucleotide BEH C18, 300 Å, 1.7 µm, 2.1 x 20 mm Column (p/n: 186011021) |
|
Column temperature: |
70 °C |
|
Sample temperature: |
6 °C |
|
Vials: |
QuanRecovery™ with MaxPeak™ HPS 12 x 32 mm Screw Neck Vials, 300 µL (p/n: 186009186) |
|
Injection volume: |
0.3 µL |
|
Flow rate: |
0.8 mL/min for the 3 min gradient and 1.5 mL/min for the 1.33 min gradient |
|
Mobile phase: |
A: 10 mM DIPEA + 100 mM HFIP in water (pH ~ 8.4) B: 10 mM DIPEA + 100 mM HFIP in MeCN/water 1/1 |
|
Syringe draw rate: |
30 µL/min |
|
Needle placement: |
1.0 mm |
|
TUV sampling rate: |
20 Hz |
|
Filter time constant: |
none |
Studying the Effect of Column Length
To study the effect of column length, input experiments were performed on a 2.1 x 20 mm column and retention model parameters were derived. Separation (chromatogram) was predicted for a 2.1 x 150 mm column and model chromatograms were compared.
The 10 mM DIPEA + 100 mM HFIP in MeCN/water phase system was used for these experiments.
Comparing Different Ion-pair Systems
The retentivity observed with different ion-pair systems was compared using the following mobile phases (Table 1).
|
Ion-pair system: |
Mobile phase composition |
|
“TEA” (triethylamine): |
A: 10 mM TEA + 100 mM HFIP in water (pH ~ 8.4) B: 10 mM TEA + 100 mM HFIP in MeCN/water 1/1 |
|
“DIPEA” (diisopropylethylamine): |
A: 10 mM DIPEA + 100 mM HFIP in water (pH ~ 8.4) B: 10 mM DIPEA + 100 mM HFIP in MeCN/water 1/1 |
|
“HA” (hexylamine): |
A: 10 mM HA + 100 mM HFIP in water (pH ~ 8.4) B: 10 mM HA + 100 mM HFIP in MeCN/water 1/1 |
|
“OA” (octylamine): |
A: 10 mM OA + 100 mM HFIP in water (pH ~ 8.4) B: 10 mM OA + 100 mM HFIP in MeCN/water 1/1 |
Table 1. The ion-pair systems used to compare the retentivity of ONs (oligo dT-mers).
"Calibration" input experiments were performed by running generic linear gradients of different gradient steepness. Retention model parameters were then derived for all phase systems and the retentivity of the four IP-systems (fraction of the two model parameters "log k0" and "S") was plotted as a function of oligonucleotide length. Please note that the ratio of logk0/S corresponds to a mobile phase composition φ*, (φ is the volume fraction of the B mobile phase in the eluent, normalized between 0 and 1) which results in a retention factor of k = 1. Since ONs follow an on-off like elution mechanism, it is reasonable to assume that a weaker mobile phase than φ* (i.e. φ < φ*) results in a very strong retention of the analytes (no migration at all) while a stronger mobile phase than φ* (i.e. φ > φ*) results in the complete release of the solutes (elution without any physico-chemical retention). Therefore, comparing the logk0/S values is an appropriate method to study and compare the absolute retentivity observed in different phase systems. For more details, please refer to referenced publications4,9,10.
Results and Discussion
The Effect of Column Length on Separation Power
It has been reported that for molecules that tend to exhibit an on-off elution mechanism, only a short inlet segment (few millimeters to 1–2 cm) of a long column bed (i.e. of a 100 or 150 mm column) participates in their effective separation.11 Therefore, long columns will not improve resolution. The steepness of the mobile phase gradient has a much more significant effect on tuning selectivity and resolution, as these molecules are very sensitive to the composition of the mobile phase.
Figure 1 shows a comparison of the expected oligo dT separations on a 2.1 x 150 mm column and an experimental result for a 2.1 x 20 mm column. The gradient time (tG) has been scaled to the column length to maintain the same intrinsic gradient steepness.
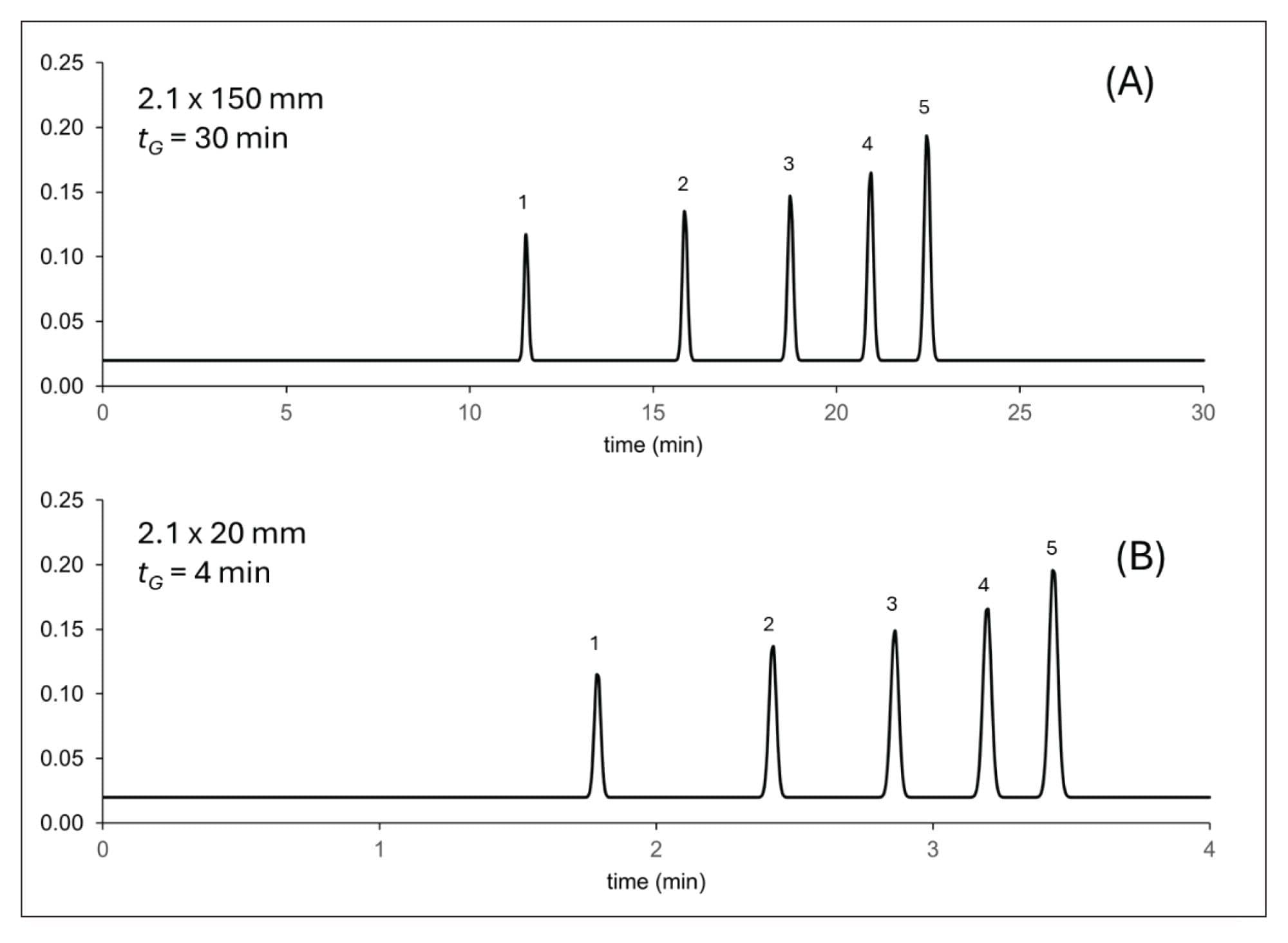
As shown in Figure 1, the two separations are nearly equivalent. The critical resolution (Rs,crit) between peaks 4 and 5 is Rs,crit = 6.1 for the 150 mm column and Rs,crit = 4.4 for the 20 mm column. However, there is a factor of 7.5 gain in analysis time. Note that the resolution with the short column can be further improved by adjusting the steepness or shape of the gradient, and the flow rate (intrinsic gradient steepness).
Experimental Comparison of Different Ion-pair Systems
Ion pairing systems are often classified as (1) weak, (2) moderate, and (3) strong. It has been shown that alkylamine retention in RPLC correlates well with the retention of paired oligonucleotides in IP-RPLC.1 Hydrophobic alkylamines act as strong IP reagents, probably due to their high affinity to the RP stationary phase.1 However, the optimal IP system depends on the analytical objective and the character of the analytes. Specific requirements such as LC-MS compatibility, length-based (size-based) or hydrophobicity-based (sequence-based) oligonucleotide separations will dictate the choice of the IP system.
It has been reported that hydrophobic (strong) IP reagents lead to improved retention of ONs due to a greater contribution from electrostatic (ion-exchange) interactions. Strong IP reagents can bind strongly to the stationary phase ligands (hydrophobic ligands) and thus the alkyl ligands of the stationary phase are masked by the charges of the adsorbed IP reagent, as the charges are likely to be directed towards the aqueous mobile phase at the top of the stationary phase-ligand-IP complex. Therefore, in IP-RPLC with a strong IP system it is reasonable to assume that the contribution of the hydrophobic retention mechanism of ONs to the overall retention is limited, and ON retention is mostly driven by ion-exchange interaction mechanism.
On the other hand, hydrophilic (weak) IP reagents are likely to favor a more hydrophobic interaction dominated retention mechanism. The stationary phase ligands are likely to be less covered by the IP reagents due to the weaker hydrophobic interaction strength (between the IP reagent and stationary phase ligands), leaving more stationary phase ligands (alkyl chains) accessible to the analytes. This is why weak IP systems have been preferred in applications where the oligonucleotides have different hydrophobicity.3,12,13
Here we compared the retention of ONs observed with two weak (TEA and DIPEA), one moderate (HA) and one strong (OA) IP agents. Figure 2 shows the retentivity of the IP-systems as a function of the oligonucleotide length for which the theory suggests a logarithmic fit because ONs can be thought of as compounds in a homologous series.7
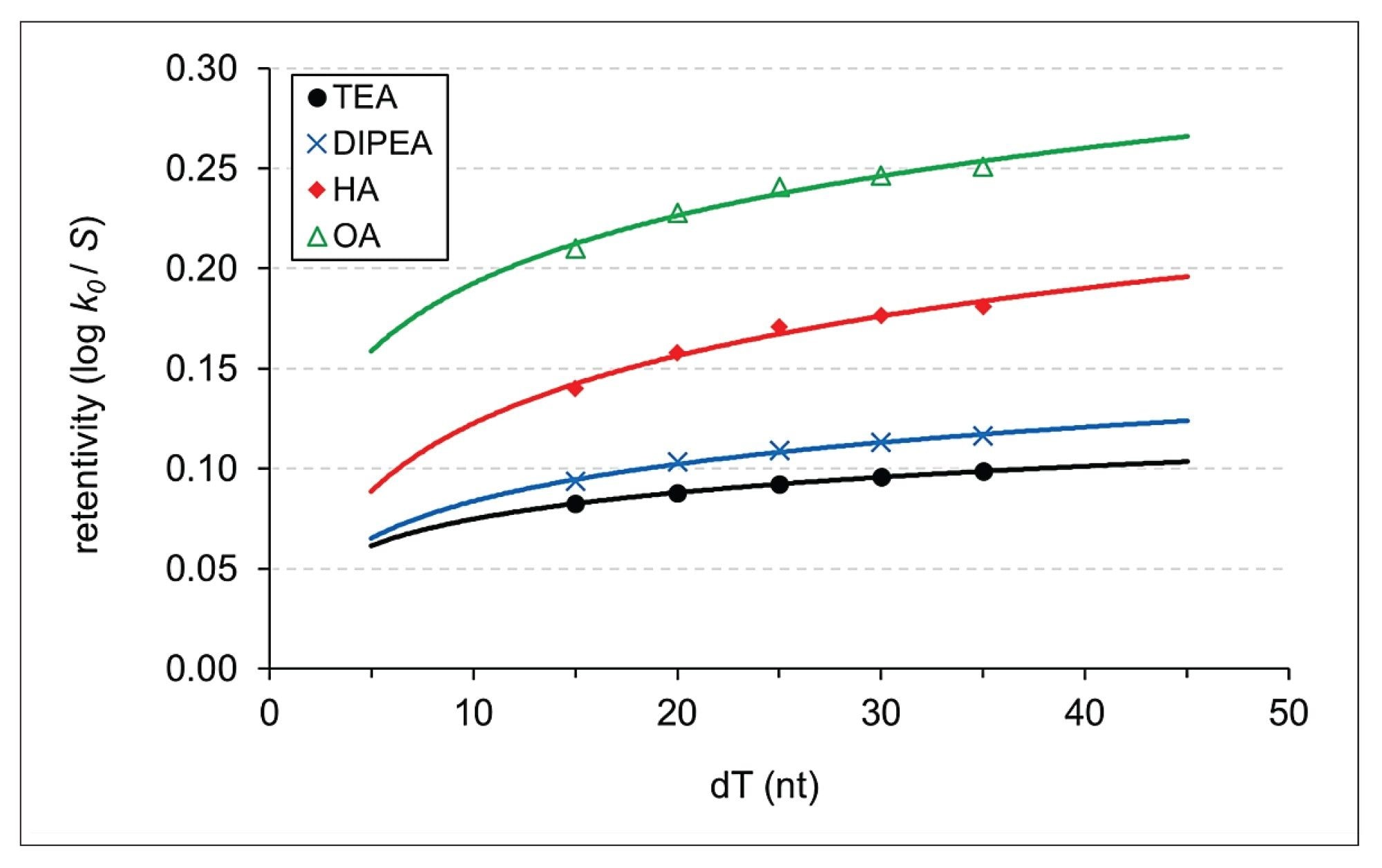
In liquid chromatography, it is an empirical observation that retention increases in logarithmic steps with increasing number of homologous units. This behavior has been reported for different series and different elution modes.14,15,16 This is often referred to as the "homologue rule". The practical consequence of the homologue rule is that higher selectivity is expected between the smaller homologues, while selectivity will decrease between the larger homologues. This tendency is independent of the nature of the IP agent. In other words, the strength of the IP system will mostly impact the absolute retention of the ON analytes but not necessarily the selectivity (relative retention). Figure 2 suggests that the elution of short ONs with weak IP agents requires much less organic co-solvent compared to strong IP agent (i.e. 8–13% MeCN with TEA and DIPEA while 20–25% MeCN with OA for the 15–35 mers).
It is up to the analyst to select the most appropriate IP system. Note, however, that strong IP agents may adsorb quasi-irreversibly onto the highly active sites of the stationary phase, requiring a more time-consuming column wash and cleaning procedure.
A Generic Platform Gradient to Maximize Selectivity
Due to the homologous behavior of ONs, the retention time of oligonucleotides will increase logarithmically rather than proportionally when running linear mobile phase gradients. As a result, the selectivity between n-1-mers will decrease significantly for larger oligonucleotides when running linear mobile phase gradients. Linear gradients can result in unnecessarily high selectivity for the short ONs and in too low selectivity for the large ONs. A more homogeneous peak distribution (retention patterns) is needed for a platform method.
If a linear mobile phase gradient results in a logarithmic (“concave”) retention time (tr) distribution for a set of compounds and the analytes elute by on-off mechanism, then setting the inverse function of tr vs φ(t) as the gradient program instead of a linear gradient, should result in a linear peak distribution (more uniform selectivity). (Producing the inverse function of the observed response curve: tr(t)=f-1(φ(t)) will result in a curve that is symmetrical to the original function.) Benefiting from the property that applying a function and then its inverse gives back the original value and knowing that originally the φ has been changed according to linear units of time, the inverse would be predicted to yield a linear response function (linear distribution in this case). Figure 3 shows the chromatographic patterns of ssDNA ladder when running linear and logarithmic mobile phase gradients.
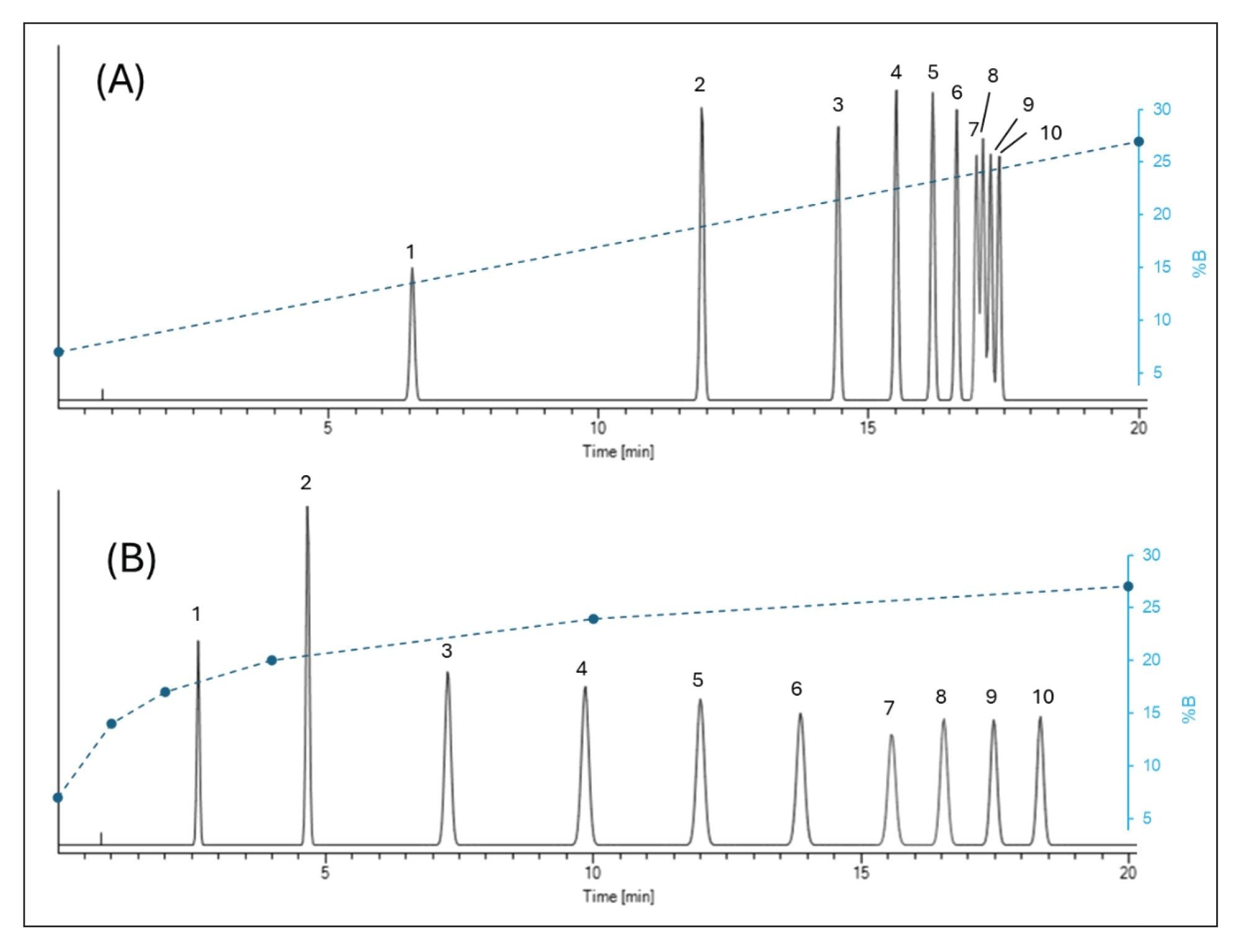
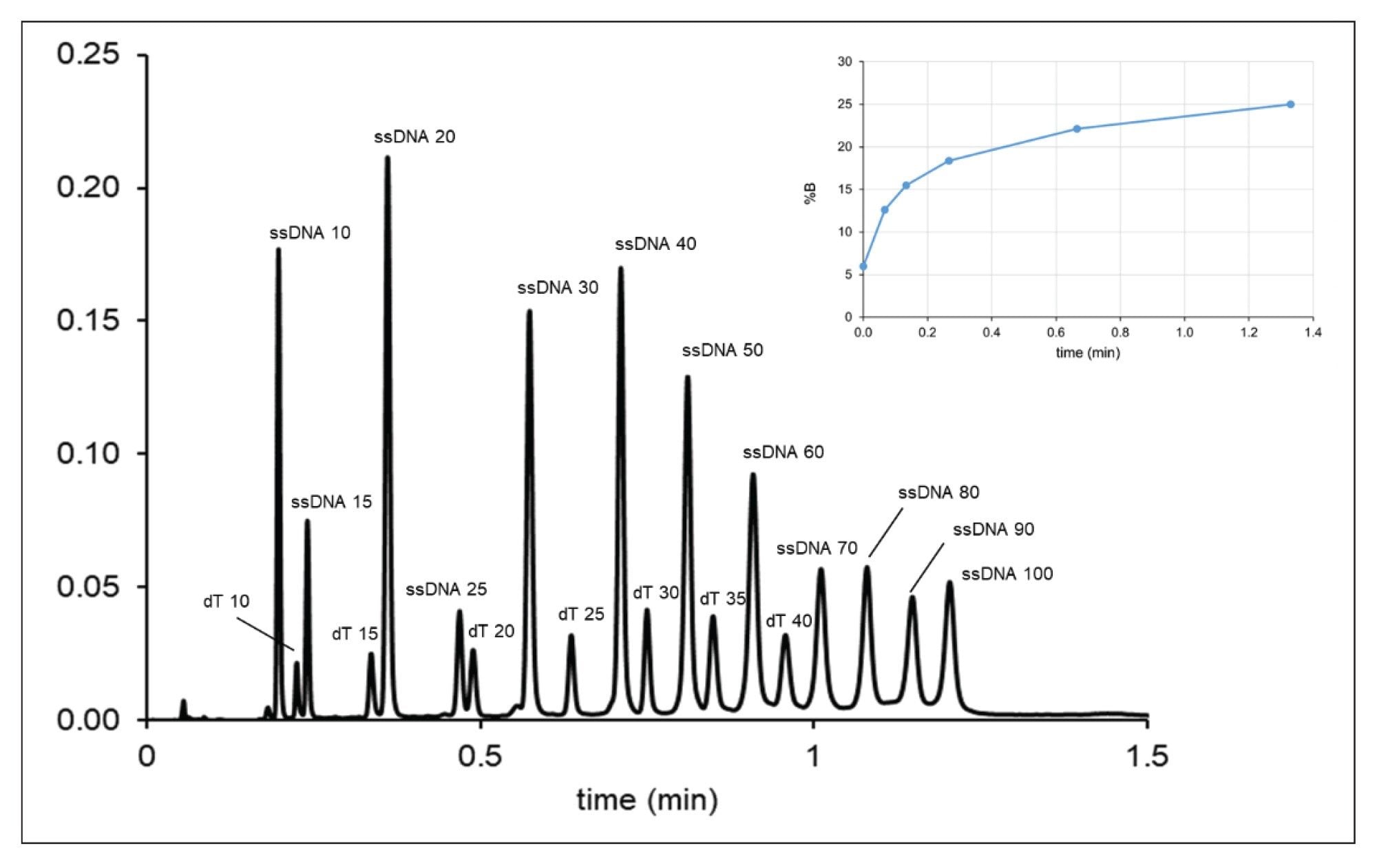
We applied this inverse gradient (logarithmic) approach for a challenging sample containing 19 compounds (various oligo dTs and ssDNAs from 10 to 100-mers, see Figure 3). Based on two input linear gradient experiments, model parameters were derived and the elution mobile phase compositions of the first peak (φfirst) and the last peak (φlast) were accurately determined. (Note that the elution composition can also be estimated from a single linear gradient run and without retention modeling, but the column dead-time and the gradient delay time must be taken into account). The starting mobile phase composition for the gradient was set to φstart = φfirst – 0.03φ, while the final mobile phase strength was set to φfinal = φlast + 0.03φ. In the example shown in Figures 4 and 5, φstart = 0.06 (6% B) and φlast = 0.25 (25% B) were set and a five-segment multi-linear (concave) gradient was programed to approach a logarithmic function. Figure 4 shows the chromatogram obtained with tG = 3 min gradient and moderate flow rate (F = 0.8 mL/min). Please note the retention pattern of ssDNAs 40, 50, 60, 70, 80, 90, and 100 mers, as they elute with nearly uniformly distributed elution distances. In contrast, when running a linear gradient, the selectivity between ssDNAs 70–100 mers will be very limited (as shown in Figure 3).
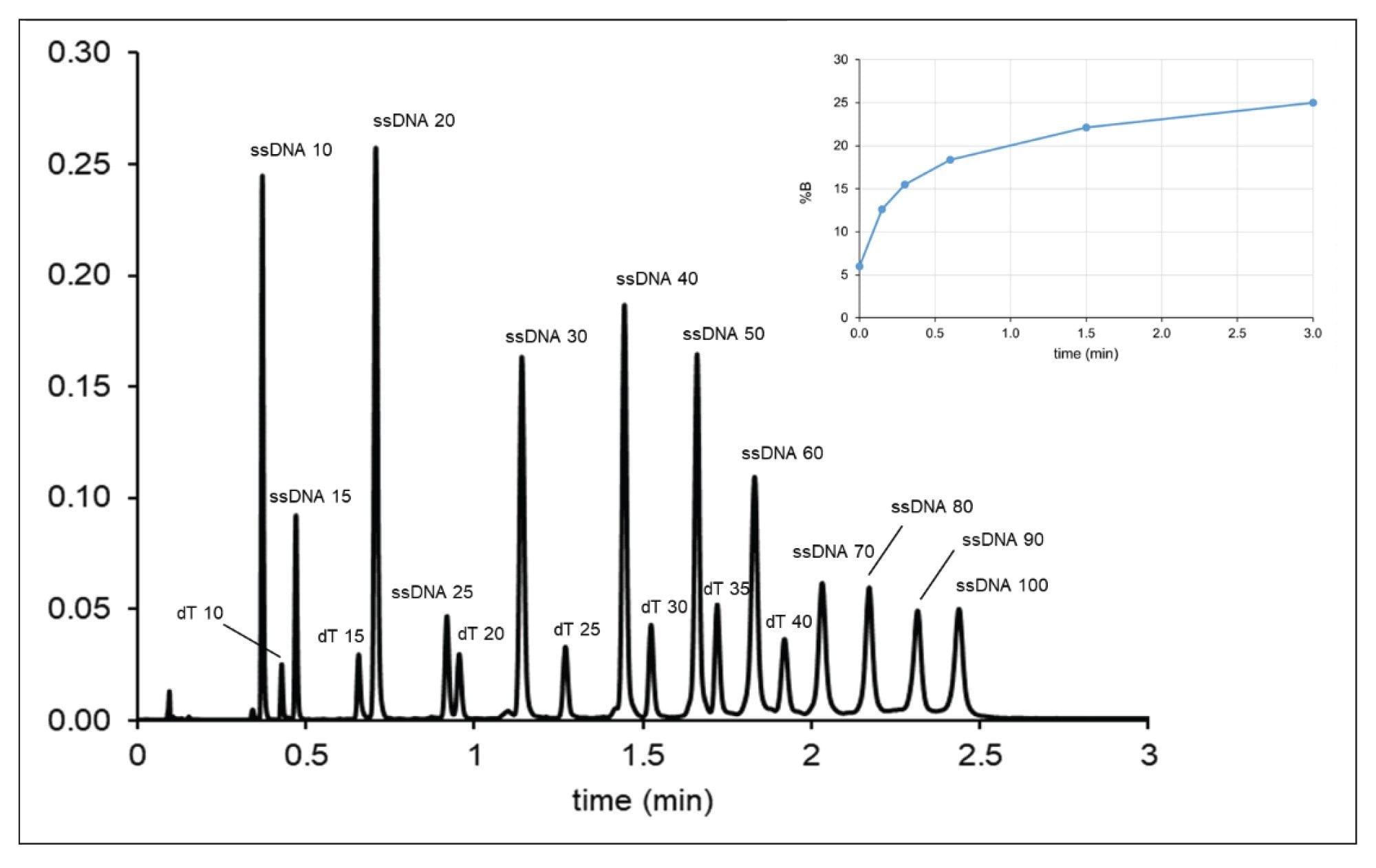
Figure 5 shows a further accelerated separation using tG = 1.33 min gradient time and high flow rate (F = 1.5 mL/min). The loss of resolution compared to chromatogram in Figure 4 is minimal. The peaks are still very sharp despite the high flow rate applied. This is due to the very strong gradient band compression effect and the high exit velocity (migration velocity at column outlet), which are consequences of the on-off elution mechanism resulting in narrow bands detected in time. The results suggest that very fast (one to three minute) generic platform gradient methods can be developed for 10 to 100-mer ON separations using ultra-short (2.1 x 20 mm) columns.
Conclusion
Oligonucleotides elute in IP-RPLC according to an "on-off" mechanism. A very small change in mobile phase strength can result in complete elution of solutes from a highly retained state (solute trapped at the column inlet). Therefore, solute retention (and thus selectivity) is only effectively controlled by a very short column segment. Hence, ultra-short columns (2.1 x 20 mm) are sufficient to effectively retain ONs.
The difficulty in method development is due to the homologous series-like behavior of ON polymers. Such behavior naturally results in decreasing selectivity with an increasing ON size. To overcome this problem, we propose here a generic (platform) logarithmic-like mobile phase gradient program that should theoretically result in the highest achievable selectivity for 10 to 100-mer ONs. By combining the logarithmic-like concave mobile phase gradient program with ultra-short columns, one-to-three-minute routine separations can be performed for mixtures containing 15–25+ compounds.
The logarithmic gradient program can be approximated by three to five segment multi-linear gradients or by the gradient curve “type 2” or “type 3” built in the Empower CDS.
Acknowledgement
The authors would like to thank Joe Fredette (Waters Corporation) for his helpful comments and discussions.
References
- M. Donegan, J.M. Nguyen, M. Gilar, Effect of ion-pairing reagent hydrophobicity on liquid chromatography and mass spectrometry analysis of oligonucleotides, J. Chromatogr. A 1666 (2022) 462860.
- M. Packer, D. Gyawali, R. Yerabolu, J. Schariter, P. White, A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems, Nat. Comm, 12:6777 (2021) 1–11.
- D.S. Levin, B.T. Shepperd, C.J. Gruenloh, Combining ion pairing agents for enhanced analysis of oligonucleotide therapeutics by reversed phase-ion pairing ultra performance liquid chromatography (UPLC), J. Chromatogr. B Anal. Technol. Biomed. Life Sci., 879 (2011) 1587–1595.
- H. Lardeux, S. Fekete, M. Lauber, V. D’Atri, D. Guillarme, High-throughput chromatographic separation of oligonucleotides: A proof of concept using ultra-short columns, Anal. Chem. 95 (2023) 10448–10456.
- Cs. Horváth, W. Melander, I. Molnár, Solvophobic interactions in liquid chromatography with nonpolar stationary phases, J. Chromatogr. 125 (1976) 129–156.
- R.R. Drager, F.E. Regnier, Application of the stoichiometric displacement model of retention to anion-exchange chromatography of nucleic acids, J. Chromatogr. A 359 (1986) 147–155.
- B. Bobály, G.M. Randazzo, S. Rudaz, D. Guillarme, S. Fekete, Optimization of non-linear gradient in hydrophobic interaction chromatography for the analytical characterization of antibody-drug conjugates, J. Chromatogr. A 1481 (2017) 82–91.
- S. Fekete, A. Murisier, M. Lauber, D. Guillarme, Empirical correction of non-linear pH gradients and a tool for application to protein ion exchange chromatography, J. Chromatogr. A 1651 (2021) 462320.
- A. Cusumano, D. Guillarme, Alain Beck, S. Fekete, Practical method development for the separation of monoclonal antibodies and antibody-drug-conjugate species in hydrophobic interaction chromatoraphy, part 2: Optimization of the phase system, J. Pharm. Biomed. Anal. 121 (2016) 161–173.
- S. Fekete, B. Bobály, J.M. Nguyen, A. Beck, J.L. Veuthey, K. Wyndham, M. Lauber, D. Guillarme, Use of Ultrashort Columns for Therapeutic Protein Separations. Part 1: Theoretical Considerations and Proof of Concept, Anal. Chem. 93 (2021) 1277–1284.
- S. Fekete, M. Lauber, Studying effective column lengths in liquid chromatography of large Biomolecules, J. Chromatogr. A 1692 (2023) 463–848.
- S.G. Roussis, M. Pearce, C. Rentel, Small alkyl amines as ion-pair reagents for the separation of positional isomers of impurities in phosphate diester oligonucleotides, J. Chromatogr. A 1594 (2019) 105–111.
- K.J. Fountain, M. Gilar, Y. Budman, J.C. Gebler, Purification of dye-labeled oligonucleotides by ion-pair reversed-phase high-performance liquid chromatography, J. Chromatogr. B Anal. Technol. Biomed. Life Sci., 783 (2003) 61–72.
- P. Jandera, Simultaneous optimization of gradient time, gradient shape and initial composition of the mobile phase in the high-performance liquid chromatography of homologous and oligomeric series, J. Chromatogr. A 845 (1999) 133–144.
- Y. Wei, C. Yao, J. Zhao, X. Geng, Influences of the mobile phase composition and temperature on the retention behavior of aromatic alcohol homologues in hydrophobic interaction chromatography, Chromatographia 55 (2002) 659–665.
- E. Peris-Garcia, M.T. Ubeda-Torres, M.J. Ruiz-Angel, M.C. Garcia-Alvarez-Coque, Effect of sodium dodecyl sulphate and Brij-35 on the analysis of sulphonamides in physiological samples using direct injection and acetonitrile gradients, Anal. Methods 8 (2016) 3941–3952.
720008460, August 2024