For forensic toxicology use only.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates to evaluate the Waters STA methodology on the ACQUITY QDa Mass Detector to facilitate screening of toxicologically relevant compounds in various biological matrices.
In the first of a two-part series on STA using the ACQUITY QDa Mass Detector, we review the strengths and weaknesses of traditional screening methods and describe a simplified screening technique for the detection of toxicologically-relevant analytes in biological matrices.
General unknown screening or systematic toxicological analysis (STA) is an essential element of the workflow applied within forensic toxicology laboratories. The main purpose of this initial screening technique is to identify putative positive samples, i.e., those containing relevant drug substances, while simultaneously eliminating negative specimens from any subsequent analytical interrogation. Typically, urine and plasma/serum are the specimens of choice.
Traditional techniques have included immunoassay (IA), gas chromatography in conjunction with mass spectrometry (GC-MS), and liquid chromatography (LC) with ultraviolet (UV) or photodiode array (PDA). Although commonly used, each of these techniques have some significant limitations, which are summarized in Table 1.
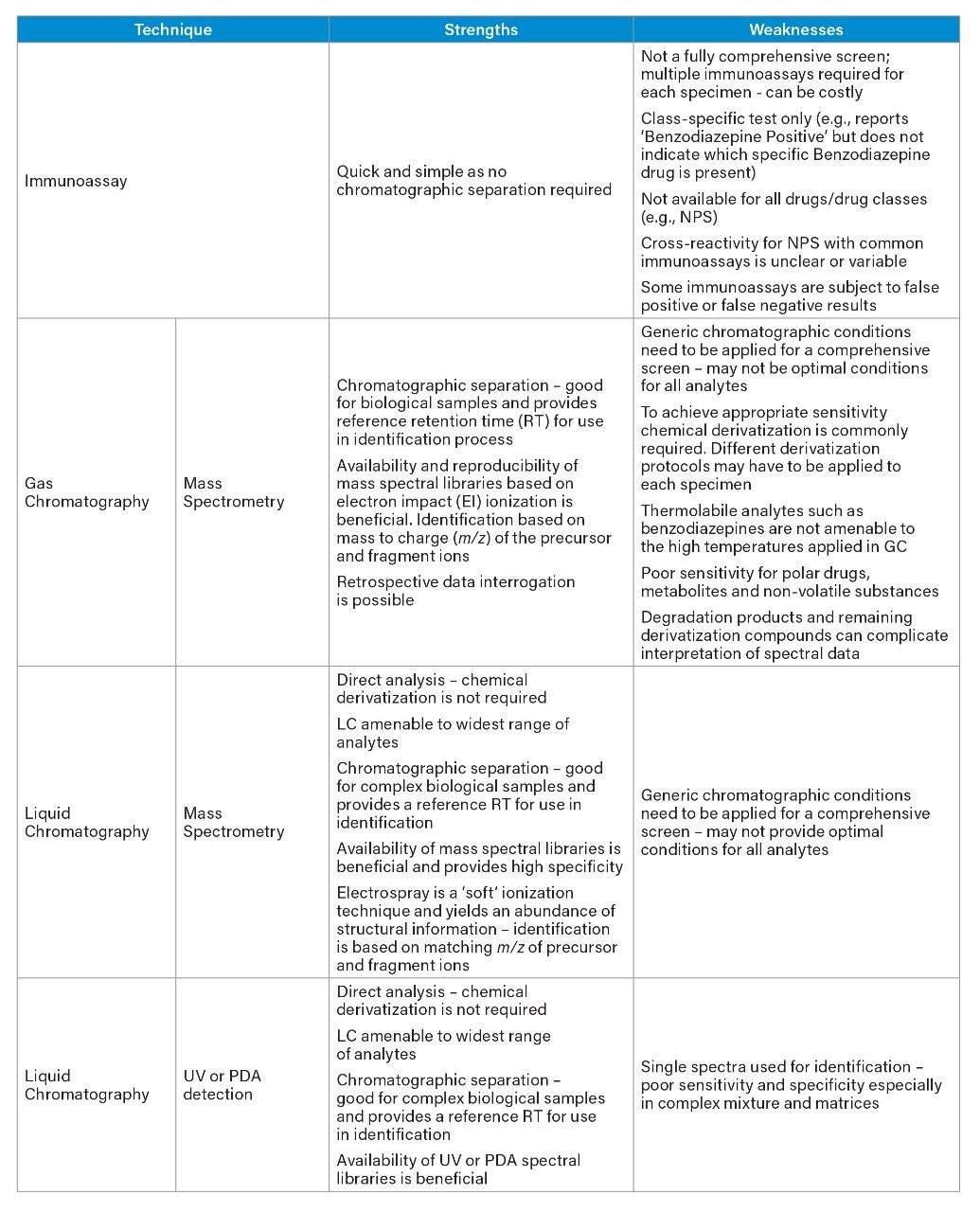
In brief, immunoassays commonly generate class-specific screening results, thus multiple kits are required to provide information for the main drug classes. In addition, assays are not available for all drugs and drug classes, therefore this approach provides a very restricted screen. False positive and false negative identifications are also a significant, and well-documented, disadvantage of immunoassays.¹
For many years, gas chromatography coupled with mass spectrometry was considered the “gold-standard” comprehensive screen and the mainstay of many toxicology laboratories. The availability of large commercial libraries based on electron impact (EI) ionization containing data for xenobiotics is certainly beneficial to the user; EI is considered to be a hard ionization technique and some analytes may be poorly detected owing to excessive fragmentation. In order to improve sensitivity, chemical derivatization is often used, which is both time-consuming and can bring additional problems. For these reasons, over the last two decades, liquid chromatographic techniques have been steadily replacing GC.
In contrast to GC, LC permits direct analysis of the drug substances without the need to chemically alter the molecule and is suitable for a wider range of drugs. Techniques based on the combination of LC and MS offer a simpler, yet more sensitive approach. Mass spectrometry detection also offers superior selectivity over PDA or UV detection and provides analyte specific detection rather than the class-specific approach of immunoassay. These advantages combine to make LC-MS the ideal tool for screening in the forensic toxicology market.
Waters first introduced the combination of LC with MS detection for STA in 2007.² Over the intervening years, this methodology evolved to reflect the advancements in technology available from Waters and users can now search against a spectral library containing 1200 compounds.3-5
Waters introduced the ACQUITY QDa Mass Detector in 2013. This instrument has already been shown to be applicable to both pharmaceutical and illicit drug identification.⁶
In this technology brief, we describe the application of the same approach to biological samples for the purposes of comprehensive toxicological screening.
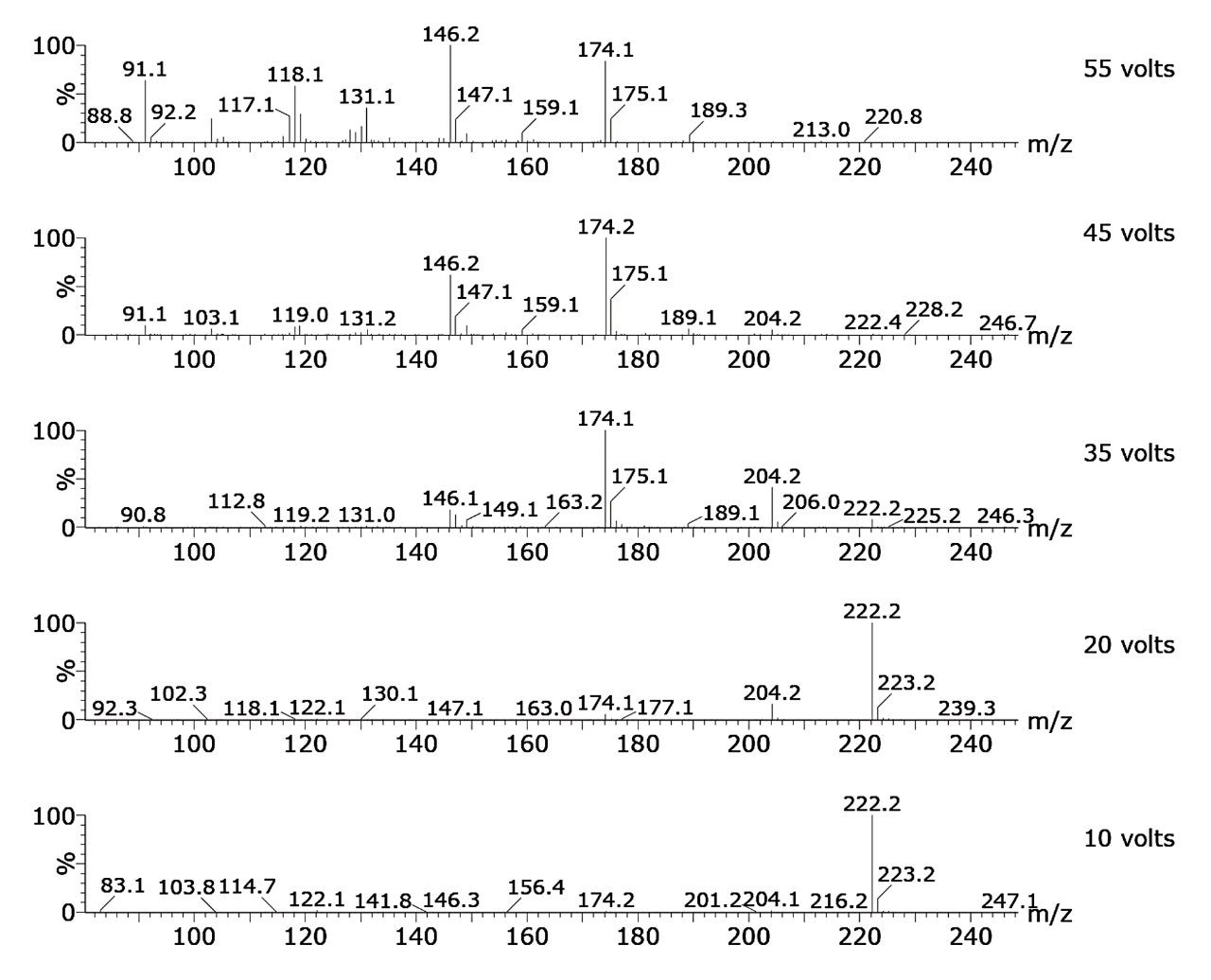
LC-MS using electrospray ionization (ESI) is ideally suited to polar, non-volatile, and thermally unstable compounds, and therefore provides a powerful means of identifying many toxicologically-relevant compounds rapidly without the need for sample derivatization, unlike GC-MS. Electrospray is a soft ionization technique that mainly leads to protonated molecules in positive mode and to deprotonated molecules in negative mode. In order to get more specific structural information, it is possible to induce fragmentation of these molecular ions in the source region of the QDa. This can be achieved by increasing the voltage applied to the sampling cone. Molecular ions then collide with neutral molecules in the source region and fragment into characteristic ions. This is referred to as in-source collision-induced dissociation (CID). Using in-source CID it is possible to generate mass spectra of different fragmentation patterns according to the value of cone voltage applied in the source (see Figure 1). Through this process, reproducible LC-MS mass spectra can be used to produce a library of mass spectra.
The ACQUITY QDa Mass Detector from Waters requires no specialist training or expertise to set up and all acquisition and processing methods used here are available free of charge from Waters Marketplace. It is the only mass detector that integrates with, and even fits on top of, your instrument stack. It uses less bench space and less energy than a traditional mass spectrometer. Cleaning and routine maintenance are minimal, thereby maximizing your uptime. It can be integrated with other Waters software platforms and detection techniques, such as UV or PDA.
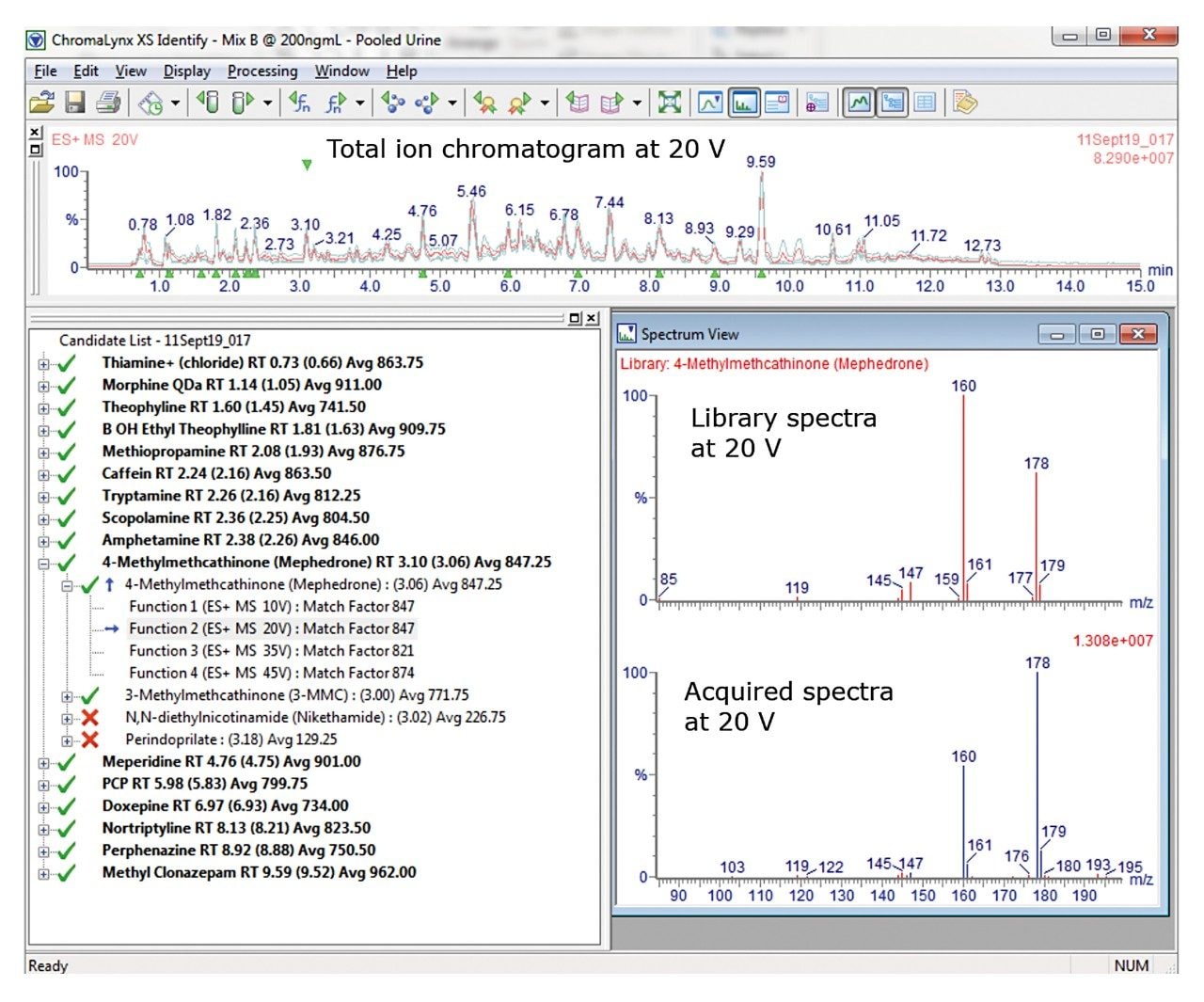
ChromaLynx is an application manager within MassLynx that allows for deconvolution of the mass spectral data and automatically processes and compares the data to the preconfigured library (containing 1200 compounds), which provides an identification through spectral library matching. The confidence with which a substance is identified is presented as an average match factor, which has a maximum value of 1000. All compounds are required to elute within 0.35 min of the retention time in the supplied library. Customers can create their own libraries from reference material or from authentic material data. Further, it is also possible to re-process previously acquired data, with updated libraries, without the need to prepare fresh samples.
One hundred toxicologically-relevant analytes were spiked into both control human urine and human plasma at 200 ng/mL and prepared using a simple Oasis PRiME HLB μElution method and analyzed using the Waters toxicology STA screening method on a Waters ACQUITY QDa Mass Detector.⁷
In this study, a positive result was achieved by having an average match factor of >700; an example of the results obtained using ChromaLynx of a mixture of compounds spiked into human urine at 200 ng/mL is shown in Figure 2.
In this technology brief a combination of liquid chromatography and ACQUITY QDa Mass Detection has been shown to provide a quick, simple, and effective way to screen for toxicologically-relevant compounds in various biological matrices. This technique offers benefits of cost savings, speed, and specificity over other established screening techniques such as immunoassay, GC-MS, and LC-UV. The same system can also be used to provide quantitative data as described by Mistry et al.⁸
720006725, January 2020