For research use only. Not for use in diagnostic procedures.
This technology brief demonstrates a high-throughput UPLC-MS/MS research method for the semi-quantitative analysis of free fatty acids in human serum samples, without the need for derivitization.
A rapid UPLC-MS/MS methodology has been developed for the research analysis of free fatty acids. This method has been demonstrated to be suitable for the analysis of physiologically relevant levels of these analytes in human serum. This method utilizes a generic LC-MS platform that can be used for various compound classes (including metabolomics, lipidomics, and proteomics), meaning it can be applied as part of a suite of analyses that are run subsequently as part of a targeted multi-omics workflow.
Fatty acids are present in biological systems as free fatty acids, as well as within more complex lipids such as triglycerides and phospholipids. Fatty acids, in various forms, make up cell membranes and are used as a concentrated energy source. The analysis of these important molecules has historically been complex and involved derivatization. Here we demonstrate a high-throughput UPLC-MS/MS research method for the semi-quantitative analysis of free fatty acids in human serum samples, without the need for derivitization. This application note is also part of a MetaboQuan-R method package.
100 µL of human serum was protein precipitated with 400 µL of methanol and centrifuged for three minutes at 25,000 g. Of the resulting supernatant, 100 µL was diluted with 100 µL of deionized water and mixed. 2 µL of this was then injected onto the UPLC-MS/MS system.
UPLC separation was performed with an ACQUITY UPLC I-Class System (fixed loop), equipped with a CORTECS T3 2.7 µm (2.1 × 30 mm) analytical column. A sample of 2 µL was injected at a flow rate of 1.3 mL/min. Mobile phase A was 0.01% formic acid (aq) containing 0.2 mM Ammonium Formate and mobile phase B was 50% isopropanol in acetonitrile containing 0.01% formic acid and 0.2 mM Ammonium Formate The free fatty acids were eluted from the column and separated with a gradient of 50–98% Mobile phase B over 1.2 minutes, followed by a 0.5 minute column wash at 98% Mobile phase B. The column was then re-equilibrated to initial conditions. The analytical column temperature was maintained at 60 °C.
Multiple Reaction Monitoring (MRM) analyses were performed using a Xevo TQ-S micro Mass Spectrometer. All experiments were performed in negative electrospray ionization (ESI-) mode. The ion source temperature and capillary voltage were kept constant and set to 150 °C and 2.0 kV respectively. The cone gas flow rate was 50 L/hr and desolvation temperature was 650 °C.
Method information was imported onto the LC-MS system using the Quanpedia functionality within MassLynx. This extendable and searchable database produces LC and MS methods as well as processing methods for use in TargetLynx for compound quantification.
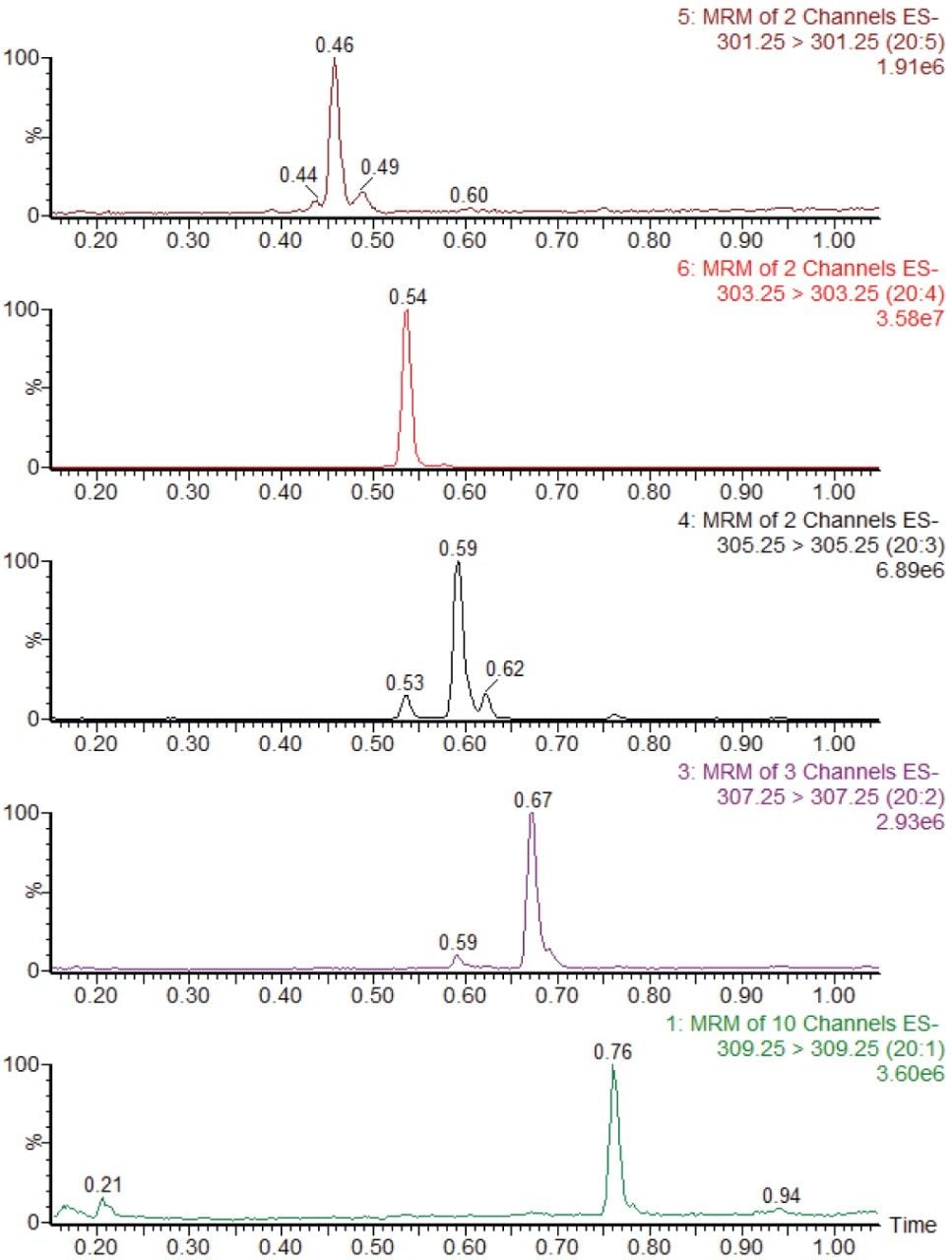
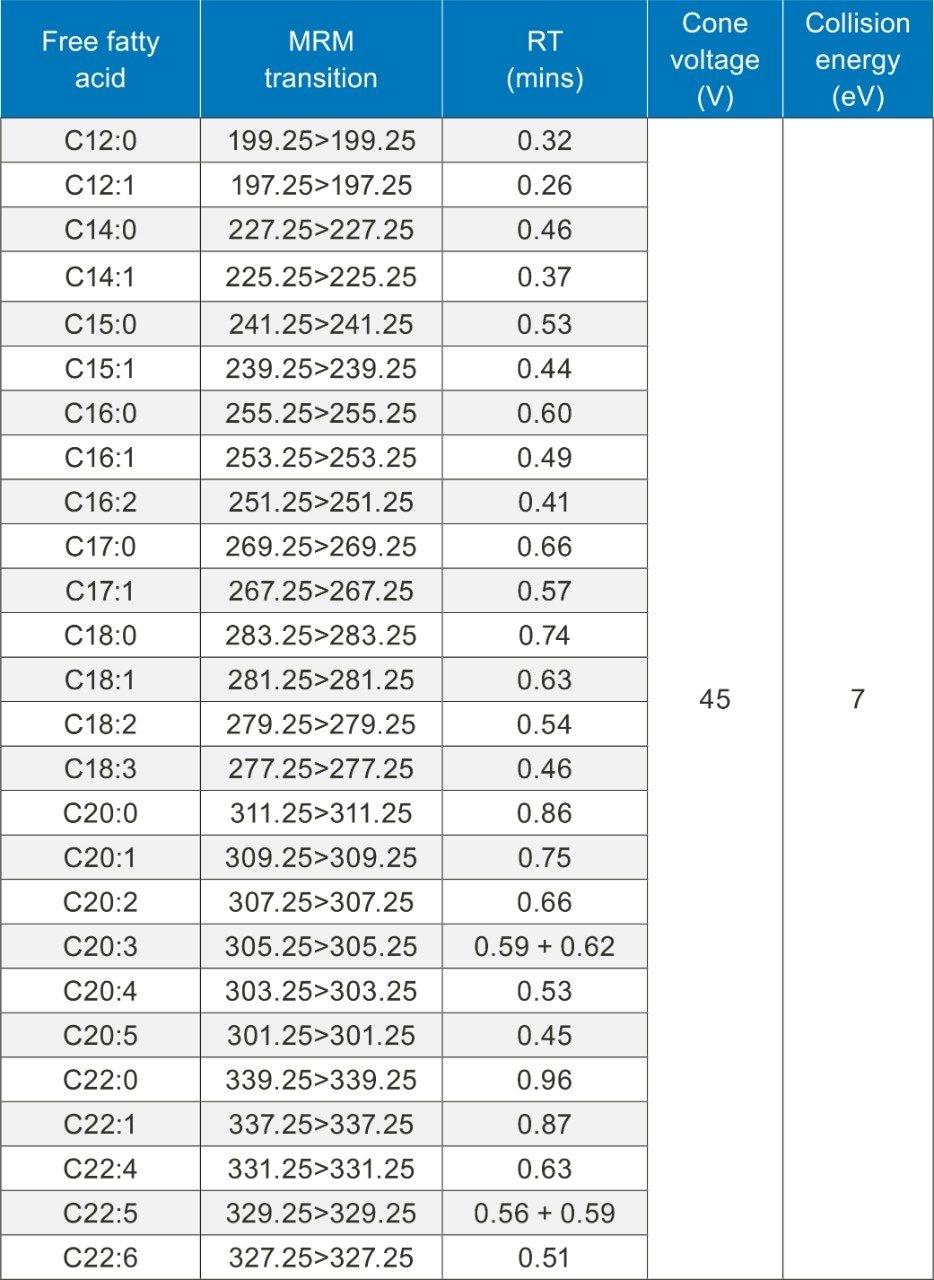
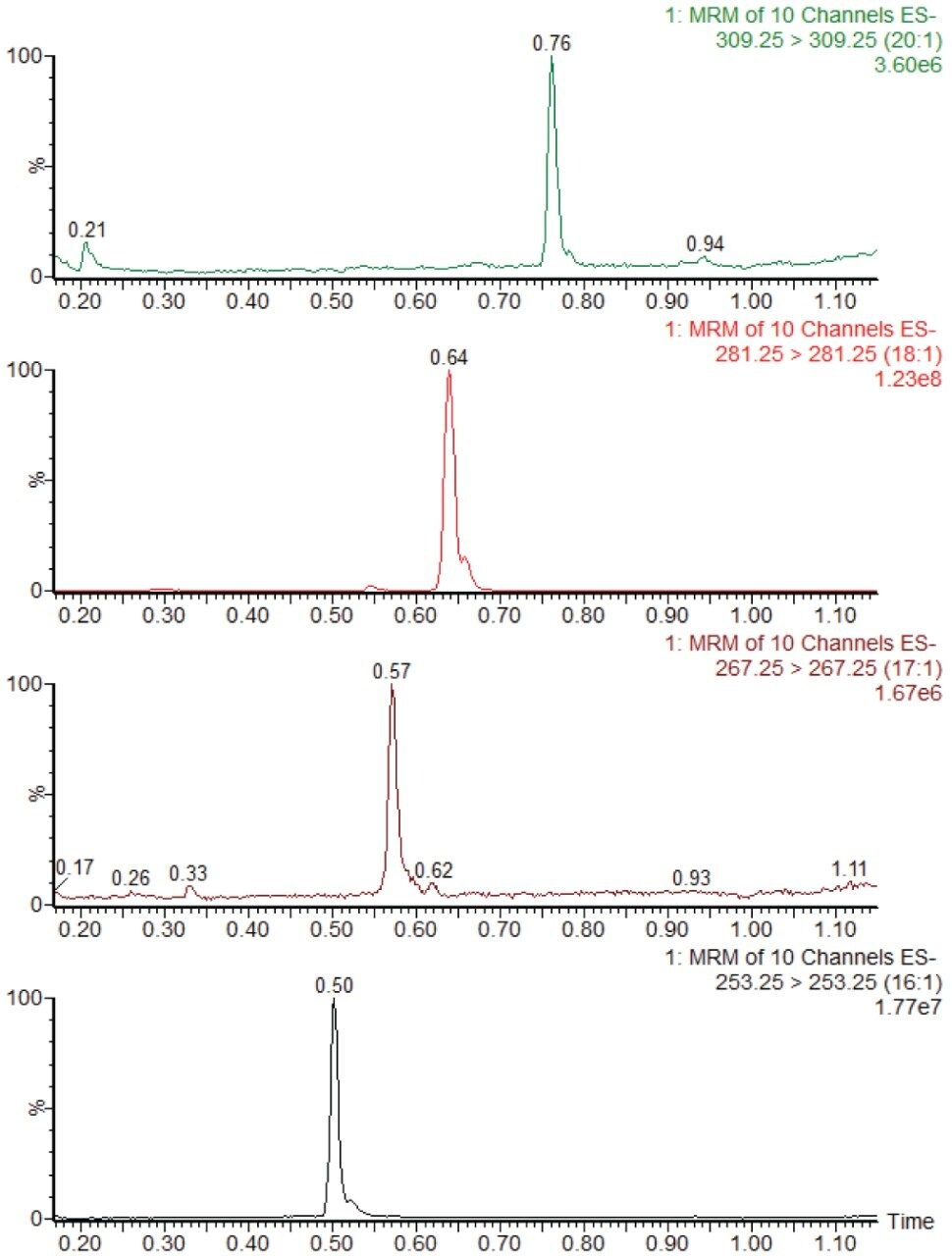
The 26 free fatty acids detailed in Table 1 were separated and detected using the LC-MS platform and extraction protocol described herein. Figure 1 shows example chromatograms for the separation achieved using the UPLC method detailed above. Fatty acids 20:3 and 22:5 have two peaks present in their MRM transitions. This is due to these compounds having two abundant positional isomers.
A rapid UPLC-MS/MS methodology has been developed for the research analysis of free fatty acids. This method has been demonstrated to be suitable for the analysis of physiologically relevant levels of these analytes in human serum. This method utilizes a generic LC-MS platform that can be used for various compound classes (including metabolomics, lipidomics, and proteomics), meaning it can be applied as part of a suite of analyses that are run subsequently as part of a targeted multi-omics workflow.
720006277, August 2018