
In this application note a sensitive and robust multiresidue method for the determination of 18 macrolide antibiotics using an ACQUITY UPLC I-Class PLUS coupled to a Xevo TQ-S micro MS/MS System.
Macrolide antibiotics, such as spiramycin, tylosin, and erythromycin are basic, lipophilic molecules that consist of 14–16-membered lactone rings, to which sugar moieties are attached. Lincosamides, such as lincomycin and pirlimycin, are structurally very different from macrolides but share a similar mechanism of action. Macrolides and lincosamides are widely approved for use in veterinary medicine to treat respiratory diseases. Additionally, macrolides are licensed for use in some countries and regions as feed additives to increase the conversion rate of feed and promote animal growth.
Although veterinary drugs play an important role in the production of livestock and poultry, incorrect use of macrolides or the shortening of the withdrawal time after treatment can possibly lead to the presence of macrolide residues in animal tissues and related foodstuffs and increases the potential risk to consumers because of allergic reactions of those sensitive to the antibiotics. To protect human health, rules exist to ensure consumer protection against the potentially harmful effects of residues in foodstuffs of animal origin (e.g., Regulation (EU) 2019/6 in the EU).1 Legislation provides for a science-based establishment of maximum residue limits (MRLs) for veterinary medicinal products. A maximum residue limit is the maximum concentration of a residue of a pharmacologically active substance that may be permitted in food of animal origin. MRLs for macrolides vary widely depending on the animal and target tissue and the country setting them.2,3 As macrolides are produced from various Streptomyces strains, they tend to be multi-component systems containing lower amounts of related compounds. For example, tylosin consists predominantly of tylosin A but with varying amounts of desmycosin (tylosin B), macrocin (tylosin C), and relomycin (tylosin D). The marker residue tends to be the most abundant component found in the tissue (e.g., tylosin A for tylosin). Since tulathromycin and most of its metabolites can be converted by acid hydrolysis to a metabolite known as CP-60,300, the EU chose this compound as the marker residue for tulathromycin, and established MRLs defined as the sum of tulathromycin and its metabolites that are converted by hydrolysis to the marker residue (CP-60,300) and expressed in tulathromycin equivalents. The inclusion of a hydrolysis step is not feasible when including multiple macrolides in one method, so here we consider this method suitable for screening tulathromycin.
In the case of a suspected non-compliant result, analysis is repeated using a validated confirmatory method, with a hydrolysis step, that complies with the residue definition. Residue monitoring plans are used to detect the illegal use or misuse of authorized veterinary medicines in food-producing animals and investigate the reasons for residue violations. In some cases, such as in the EU, exporting countries must also implement a residue monitoring plan that guarantees an equivalent level of food safety.
Food business operators also undertake chemical analyses to check for the presence of residues in tissues of animals within their supply chain for due diligence and positive release purposes. In addition to checking MRL compliance, there is growing concern about antibiotic resistance and its threat to human health. Acquired resistance to macrolides and lincosamides among food animal pathogens, including some zoonotic bacteria, has now emerged.4
Therefore, it is important to develop simple but accurate methods for the determination of residues of antibiotics in animal tissues. This application note describes the validation of a method for the determination of 18 macrolide antibiotic veterinary drugs in bovine muscle tissue using the Waters ACQUITY UPLC I-Class PLUS coupled to the Xevo TQ-S micro.
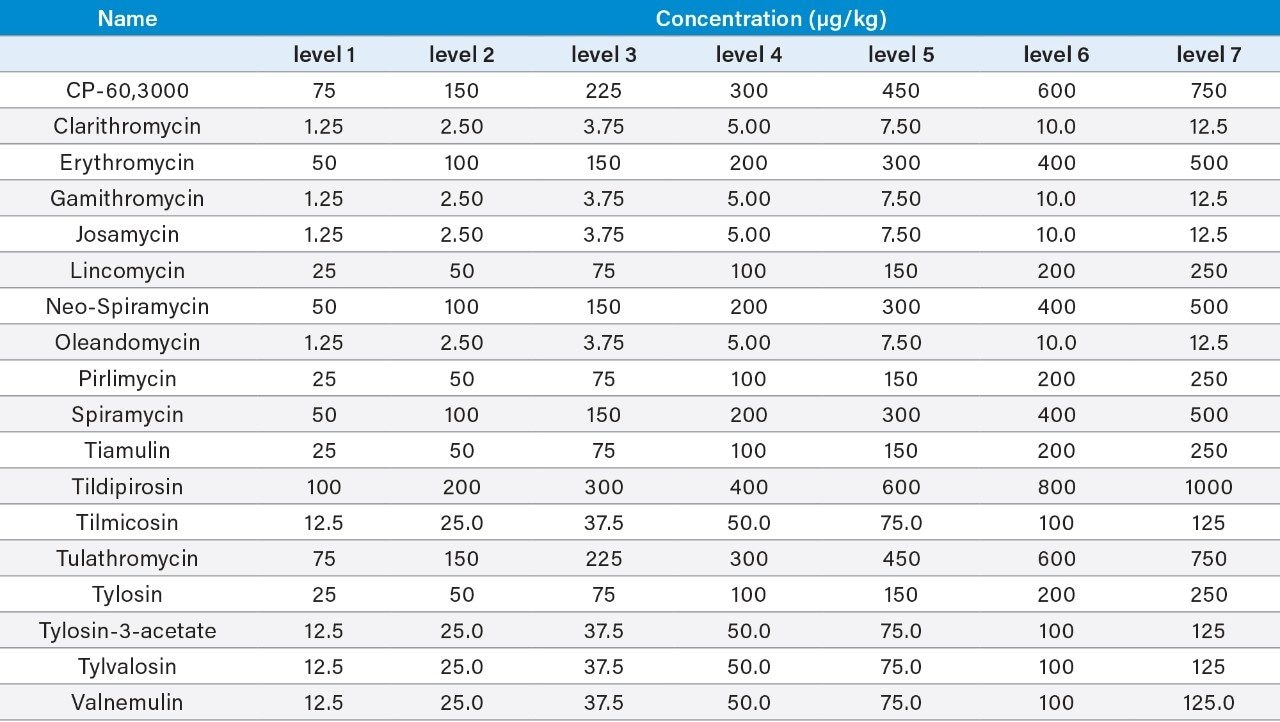
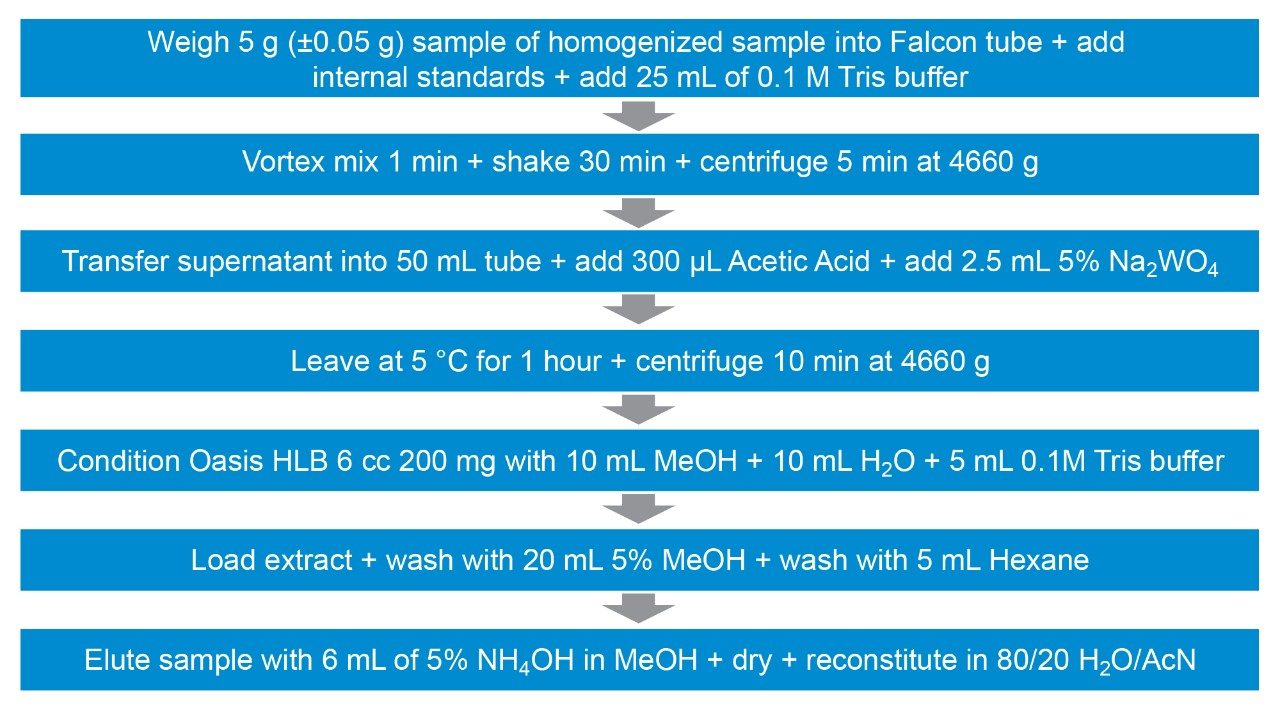
Bovine muscle tissue was extracted, after the addition of internal standards, using a liquid extraction followed by SPE clean-up (see Figure 1 for more detail). Matrix-matched standards were prepared in bovine muscle tissue extract, previously shown to be blank, at the concentrations shown in Table 1.
|
System: |
ACQUITY UPLC I-Class PLUS with FTN Sample Manager |
|
Column: |
ACQUITY HSS T3 2.1 × 100 mm (p/n: 186003539) |
|
Column temp.: |
50 °C |
|
Sample temp.: |
10 °C |
|
Injection parameters: |
1 μL |
|
Mobile phase A: |
Water + 0.1% formic acid |
|
Mobile phase B: |
Acetonitrile + 0.1% formic acid |
|
Sample manager wash: |
Methanol |
|
Time |
Flow rate (mL/min) |
%A |
%B |
Curve |
|---|---|---|---|---|
|
0.00 |
0.4 |
90 |
10 |
— |
|
0.50 |
0.4 |
90 |
10 |
6 |
|
7.50 |
0.4 |
43 |
57 |
6 |
|
9.00 |
0.5 |
0 |
100 |
1 |
|
10.00 |
0.4 |
90 |
10 |
1 |
|
MS system: |
Xevo TQ-S micro |
|
Polarity: |
ES+ |
|
Capillary voltage: |
1.0 kV |
|
Source temp.: |
150 °C |
|
Desolvation temp.: |
500 °C |
|
Desolvation gas flow: |
1000 L/hr |
|
Cone gas flow: |
50 L/hr |
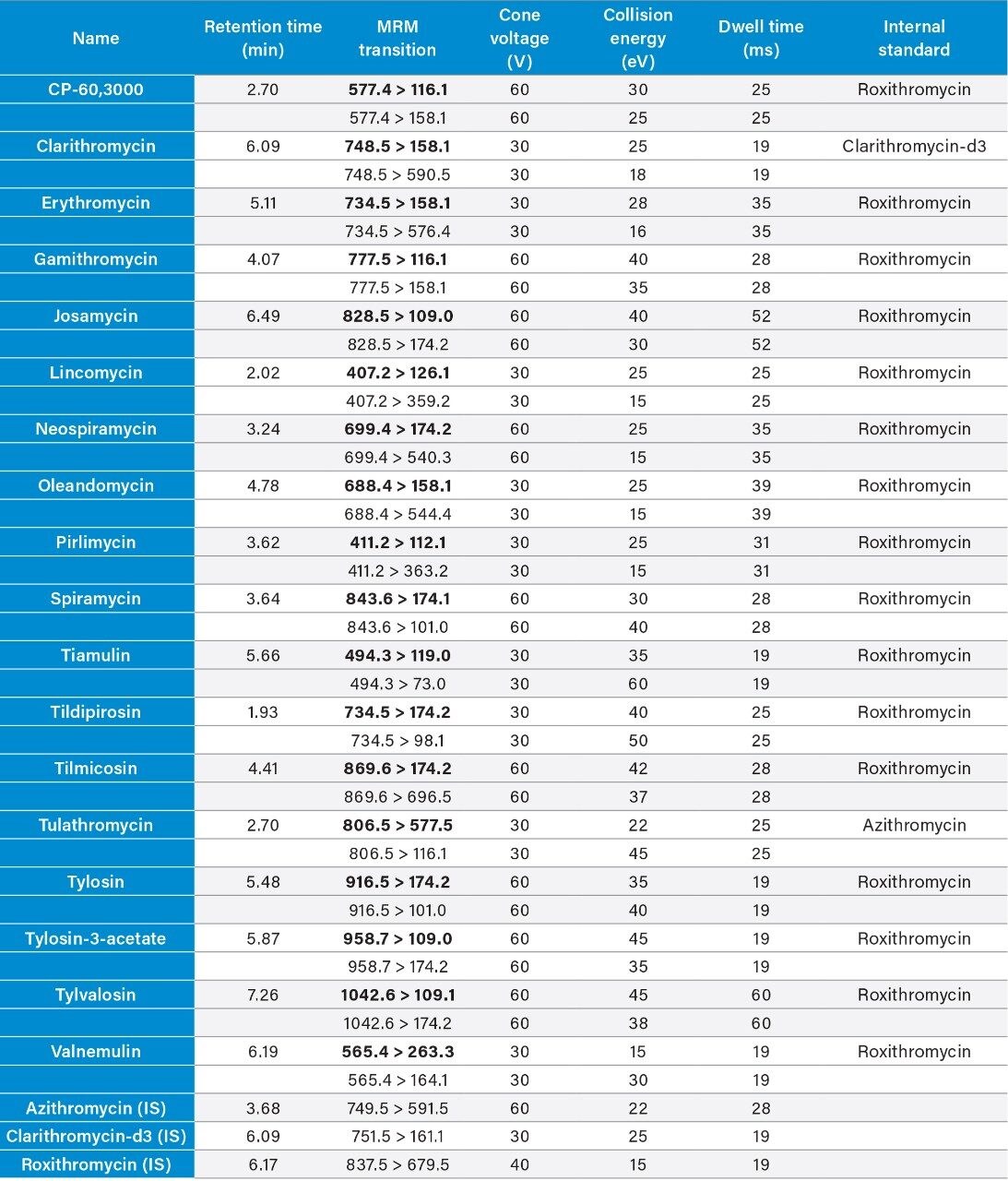
Two MRM transitions per compound were used. The dwell times were set automatically using the autodwell function to give a minimum of 12 data points across each peak. The data were acquired using MassLynx Software and processed using TargetLynx XS Application Manager. Table 2 summarizes the MRM transitions and the actual dwell time settings. The quantification traces are noted in bold.
Validation was performed following the Commission Decision 2002/657/EC guidelines.5 The following parameters were assessed: identification, selectivity, linearity, trueness (expressed as recovery), within-laboratory repeatability (RSDr), within-laboratory reproducibility (RSDRL), decision limit (CCα), and detection capability (CCb). Identification was assessed by examining retention times, ion ratios, and identification points. The selectivity of the method was investigated through injecting standard solutions of all analytes and internal standards individually and through testing bovine muscles from different animals, to check the presence of any interferences eluting at and around the retention times of the analytes. The linearity of the curves and individual residuals were checked. The trueness, RSDr, and RSDRL were derived from data from the replicate spiked samples, performed on three separate days by the same analyst. For MRL substances, parameters were assessed at 0.5, 1, and 1.5 times the MRLs established by current legislation. For those compounds with no EU MRL, assessment was made at 0.5, 1, and 1.5 times a target level (TL). CCα and CCb were calculated from the RSDRL, as defined in 2002/657/EC.
The HSS T3 column provided excellent retention and peak shape for all the analytes (Figure 2). All peaks eluted between 1.9 and 7.3 minutes with a total run time of 11 minutes.
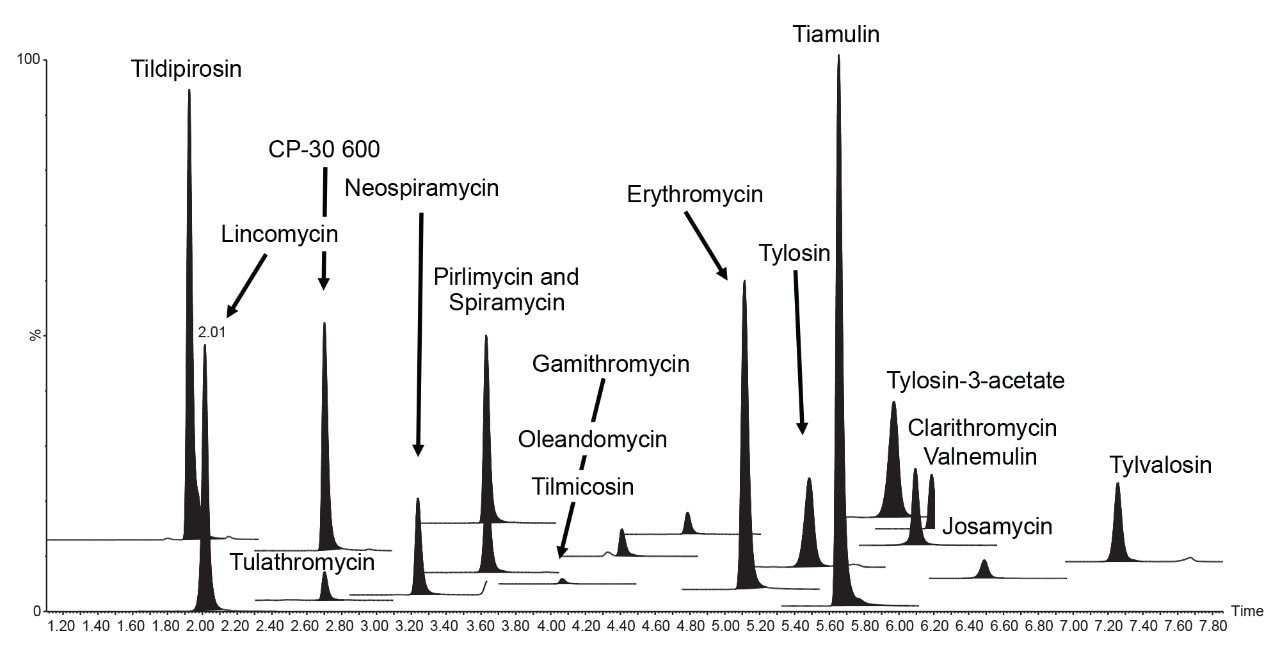
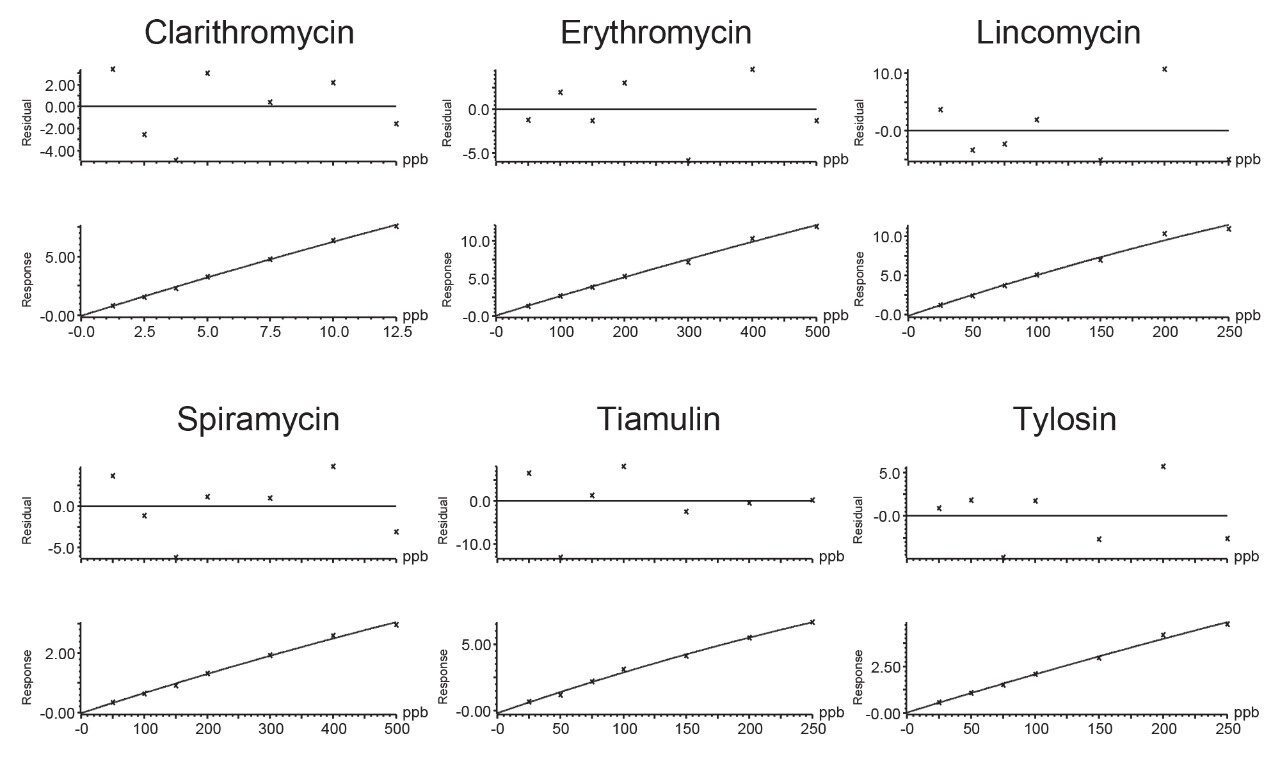
The specificity was good, as seven blank samples were prepared and analyzed on each of the three days and no significant interferences were found in the region of expected elution of the target analytes. Traces of clarithromycin (ca. 0.1 μg/kg), pirlimycin (ca. 0.7 μg/kg) and tylvalosin (ca. 0.3 μg/kg) were detected but at concentrations much lower than the MRLs. The two transitions for each analyte, enough to meet the required identification points (three for MRL substances and four for banned substances), gave peaks with ion ratios and retention times within the recommended tolerances, when compared with the standards. A seven-point calibration curve was prepared in matrix extract and acquired on each day. Quadratic fit with 1/X fit weighing was applied and values for correlation of determination (R2) of the matrix validation curves were almost all >0.99, with individual residuals <20%, demonstrating reliable quantification of all the macrolides. The calibration graph for tylvalosin from day 1 had a R2 value of 0.98. Some examples of typical calibration curves are given in Figure 3.
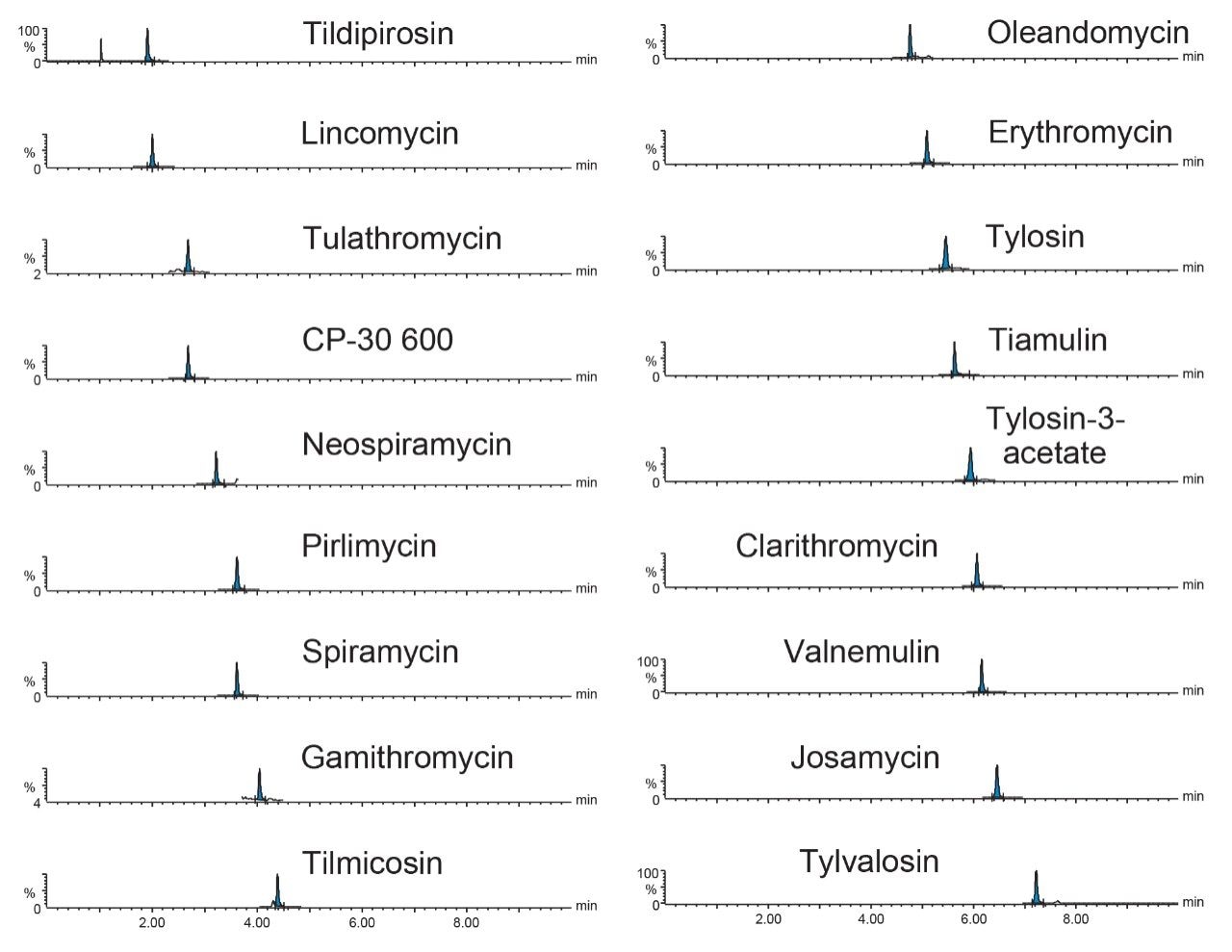
Excellent sensitivity was demonstrated from the analysis of matrix-matchedstandards. Figure 4 shows typical chromatograms for a selection of the macrolides from the analysis of the matrix-matched standard at the lowest concentration, which indicates that the method is capable of detection of macrolides in extracts at much lower concentrations. This also provides options for further dilution of the final extracts to mitigate matrix effects and to reduce further the potential contamination of the system.
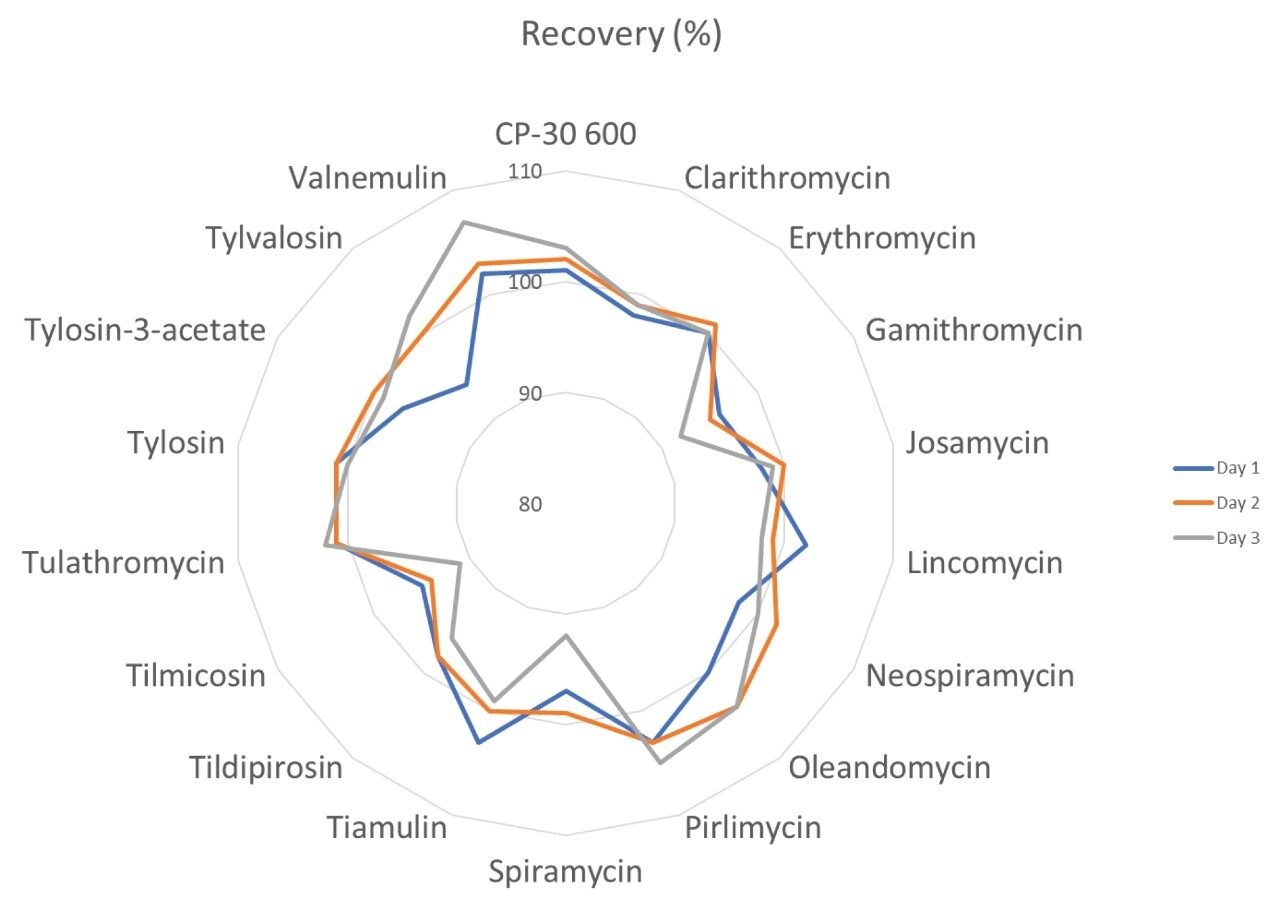
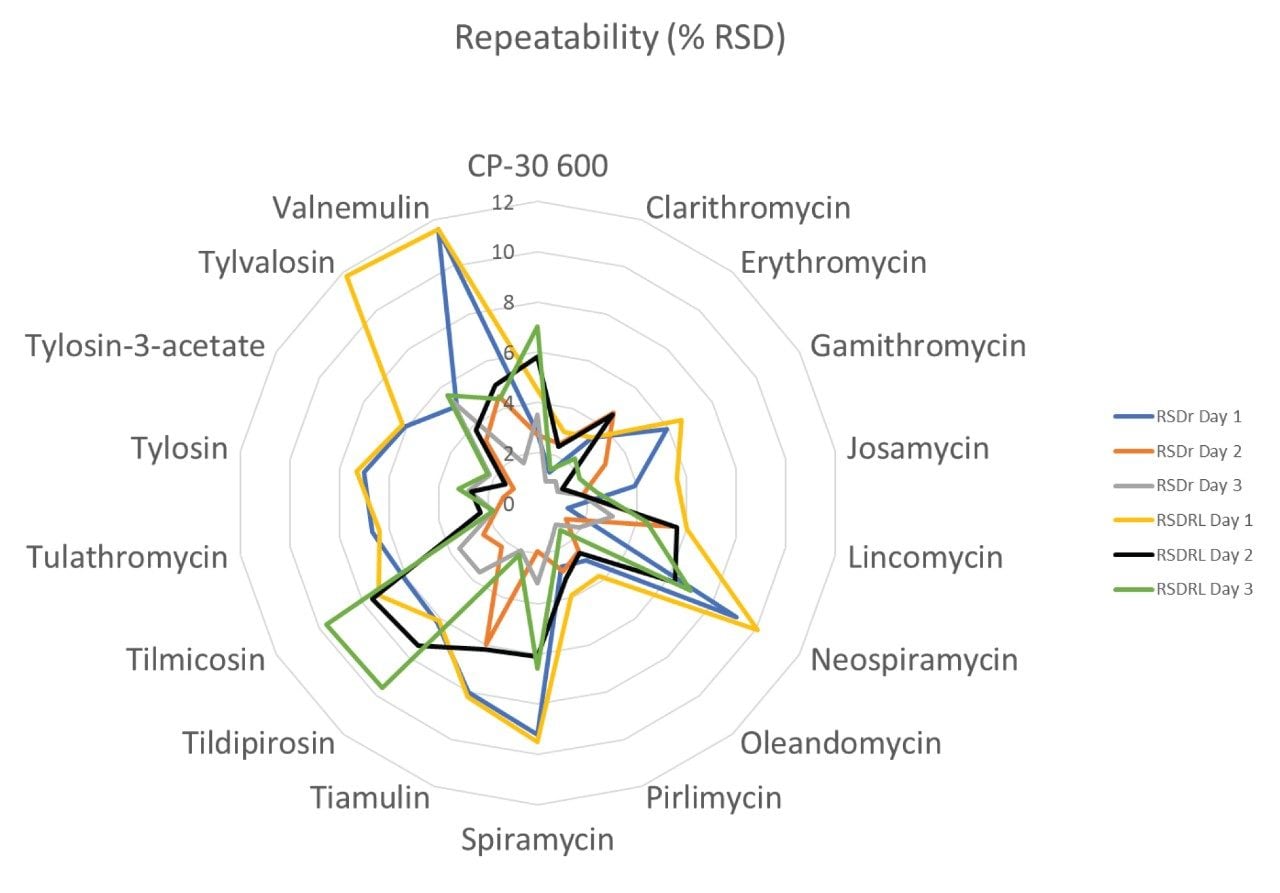
The trueness, expressed by measured recovery, was evaluated using the data from the analysis of the spiked samples over the three days. The mean recoveries for each set of seven spikes, at the three concentrations, prepared and analysed over three days, were within the range 81–111%. All but CP-60,3000 gave acceptable values for trueness. CP-60,3000 was quantified using roxithromycin as an internal standard, which may have contributed to its marginal failure (95–111%; i.e., outside the 110% limit). The repeatability of the method was satisfactory for all analytes in both RSDr (0.9–11.6%) and RSDRL (1.4–11.6%) studies. Trueness and repeatability are shown in Figures 5 and 6 and in Table 3, which also provides values for CCα and CCβ.
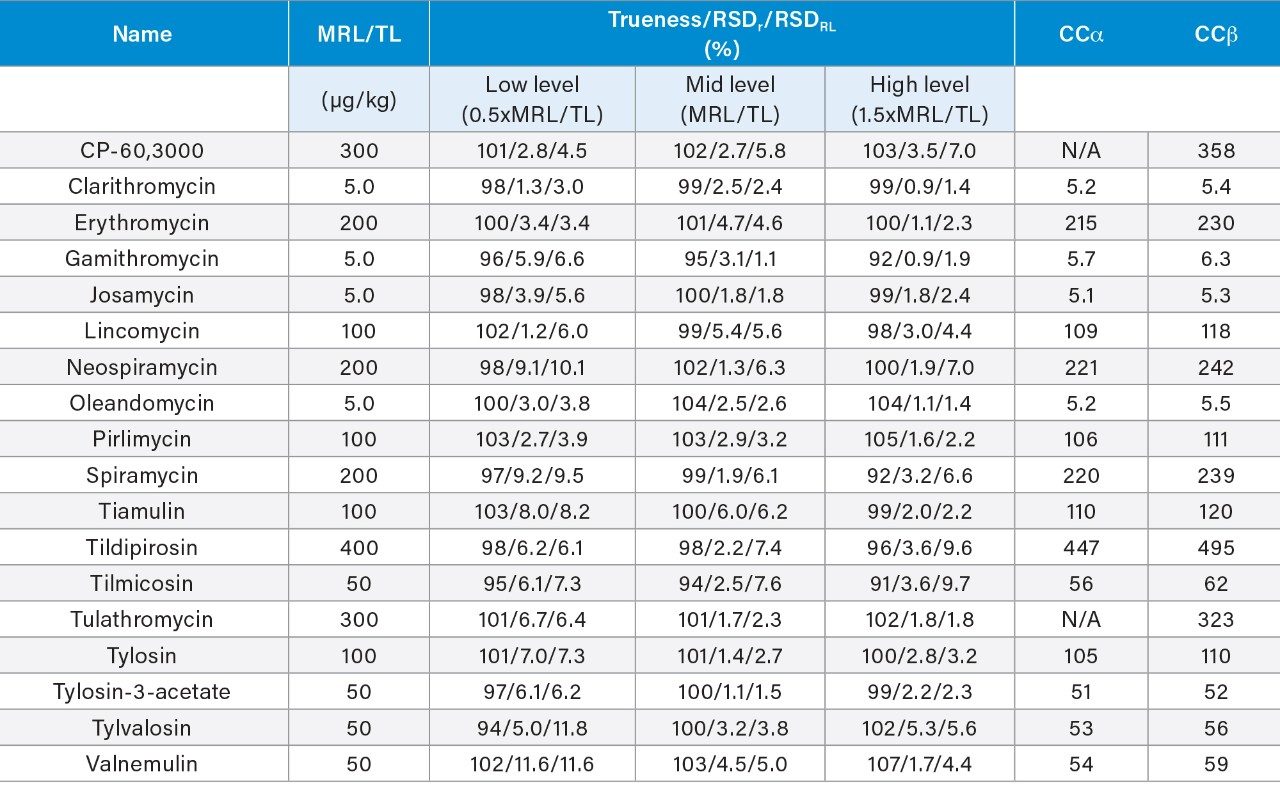
The method described here proved to be a sensitive and robust multiresidue method for the determination of 18 macrolide antibiotics using an ACQUITY UPLC I-Class PLUS coupled to a Xevo TQ-S micro MS/MS System. The method allows for a fast and reliable quantitation down to concentrations well below typical MRLs and was successfully validated according the European Commission Decision 2002/657, presenting satisfactory results for all macrolides in bovine muscle tissue. The procedure can also be applied to other animal and fish tissues after suitable validation.
720006750, February 2020