For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
The targeted quantitative analysis of protein mixtures requires proteolytic digestion coupled with a strategy that can accurately quantify proteins with high specificity, sensitivity, dynamic range, reproducibility, and robustness. The Xevo TQ-S System has seen substantial improvements in instrument performance, enabling highly accurate and sensitive protein quantification, using stable isotope peptides as internal standards, which will be presented here.
Detect and quantify potential protein biomarkers with maximum specificity and sensitivity using Xevo TQ-S.
Multiplexed multiple reaction monitoring (MRM)/selected reaction monitoring (SRM)- based quantification assays are an attractive alternative to biochemical-based protein quantification methods, such as ELISA, in terms of speed and cost savings during the early and late validation/verification stages of protein biomarkers. The requirements for such studies place a demand on the performance of the analytical LC-MS system, requiring the highest sensitivity, specificity, speed, and robustness possible to quantify the proteins of interest present within a sample. As such, technology improvements are indispensable. The use of Xevo TQ-S and TRIZAIC UPLC systems and the results for the quantitative analysis of pre-eclampsia related calcyclin peptides in formalin-fixed paraffin embedded placenta are described.
The suitability of AQUA stable isotopes and chemically equivalent internal standards for protein quantification in solution, synthetic matrix, and formalinfixed paraffin tissue samples was investigated. Limit of detection (LOD) studies were conducted by means of experiments where calcyclin peptides with an extra glycine inserted in the middle of the amino acid sequence were spiked into a synthetic matrix. MRM assay linearity and protein concentrations were determined with AQUA stable isotope peptides as the internal standard. The peptides were separated and quantified using a Waters TRIZAIC UPLC System with nanoTile Technology coupled with a Xevo TQ-S Mass Spectrometer. The data were acquired in time-scheduled MRM mode of acquisition. Quantification was conducted with TargetLynx Application Manager.
Figure 1 shows the product ion spectra of one of the calcyclin peptide candidates for MRM quantification and two possible internal standard equivalents. The chemically synthesized glycine inserted equivalent illustrates similar ionization, fragmentation, and chromatographic characteristics as the native peptides of interest. It was spiked together with native standards into a synthetic matrix, shown in Figure 2, to access the limits of detection of the MRM assay, suggesting that low attomole on-column quantification should be achievable.
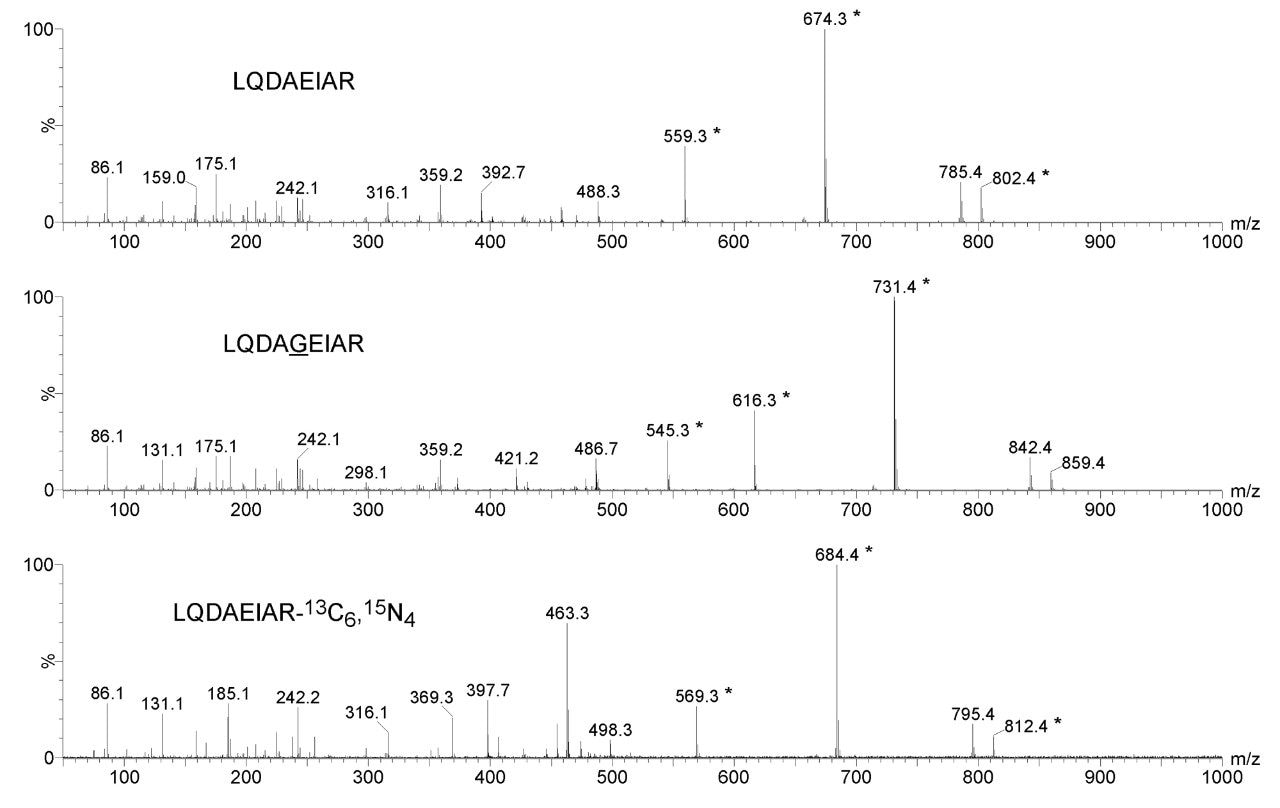
Figure 1. Product ion spectra of LQDAEIAR, glycine inserted chemical equivalent LQDAGEIAR and AQUA standard LQDAEIAR-13C6,15N4.
* Fragment ion candidates for MRM quantification.
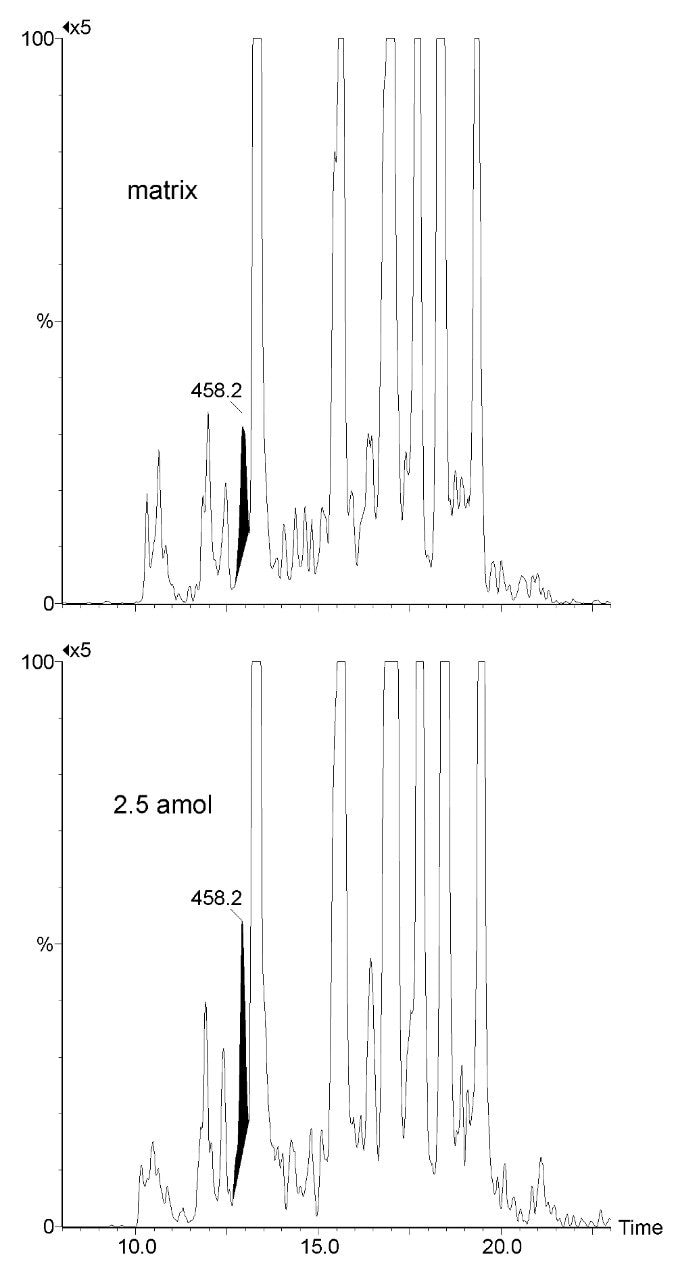
Figure 3 illustrates an MRM quantification curve for one of the AQUA stable isotope peptides, covering at least four orders of linear quantification dynamic range. Enzymatic digests of approximately 300 microdissected trophoblast and stroma cells were spiked with two stable isotope AQUA peptides in order to determine calcyclin levels. The MRM channels of one of the native peptides and the corresponding AQUA stable isotope internal standards are shown inset of Figure 3. The determined amounts were approximately 0.3 and 7.0 attomole per trophoblast and stroma cell, respectively, corroborating previously reported results.1
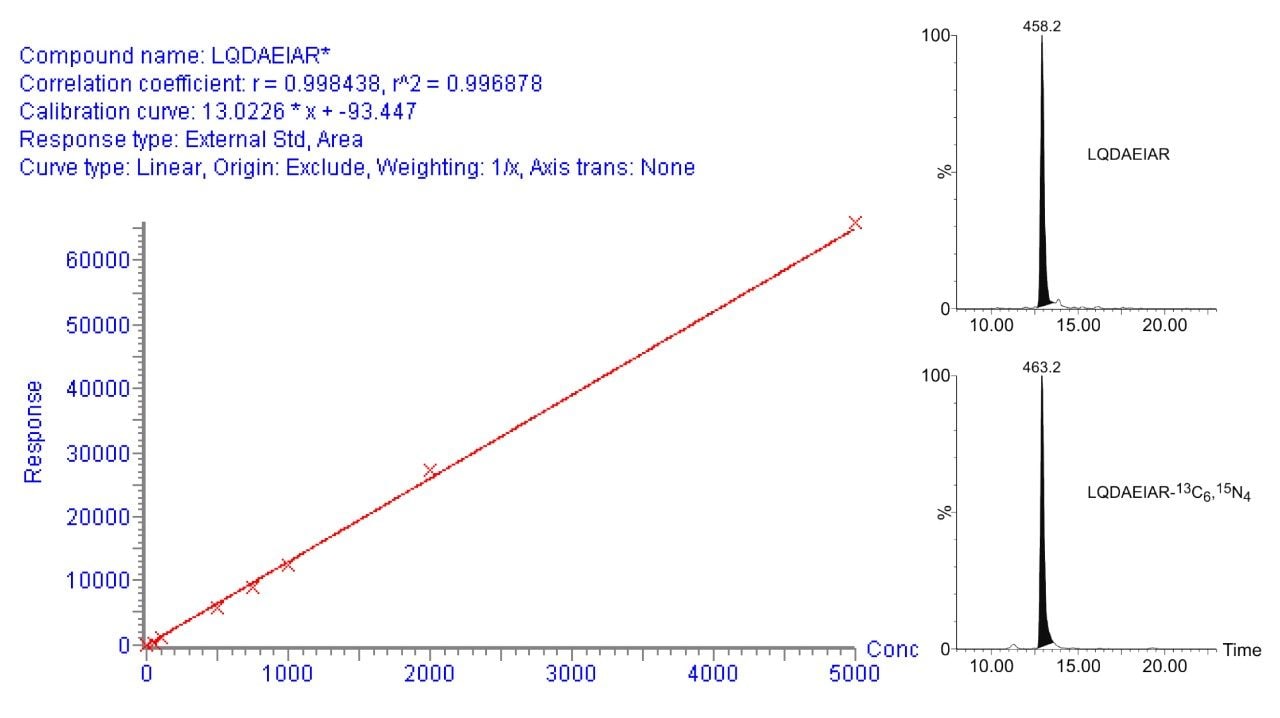
The absolute quantification of clusterin – a potential biomarker for pre-eclamsia formalin-fixed paraffin embedded placenta using AQUA stable isotopes and chemically equivalent peptides has been illustrated. The additional sensitivity of the Xevo TQ-S affords the detection and quantification of calcyclin in a smaller number of microdissected placental cells.1 The MRM-based validation of clusterin in plasma on a larger patient cohort is currently under study.
720004155, November 2011