
This application note demonstrates the capability of multi-dimensional liquid chromatography for the use of mass detection with an unmodified USP monograph for lidocaine and prilocaine cream to confirm identity of an impurity peak.
U.S. Pharmacopeia (USP) compendial methods are routinely adopted by pharmaceutical companies during the development and quality control of drug substances and finished drug products. Often, these methods use mobile phases with non-volatile buffers, which are not suitable for mass spectrometry (MS). In some cases, mass spectrometry will enable quick and accurate identification of new or unknown components that may develop during the formulation process or routine testing. Incorrect identification or failure to identify these components may compromise the safety and efficacy of the pharmaceutical product.
Changes to the USP monographs or registered methods to make them compatible with mass spectrometry will require a re-validation. In order to obtain mass spectral information for a sample tested using a non-MS compatible method, the sample must typically be isolated using a preparative LC system, purified, and reanalyzed under MS conditions.1 This process can be time consuming, and require additional instrumentation and development work. A multi-dimensional liquid chromatography with heart-cutting technique enables use of mass detection directly with the unmodified methods. With this system, peak of interest is heart-cut from the first dimension (1D) column eluted with non-MS compatible method and transferred into a holding loop. The heart-cut volume is then refocused onto a trap column and transferred to the second dimension (2D) separation column, which uses MS friendly conditions. This approach allows quick identification of unknown components by mass detection, eliminating the need to modify the method.
In this work, we demonstrate the capability of multi-dimensional liquid chromatography for the use of mass detection with an unmodified USP monograph for lidocaine and prilocaine cream2 to confirm identity of an impurity peak. An impurity peak observed in the assay preparation solution is heart-cut, trapped, and transferred to the second dimension for analysis by mass detection. The heart-cutting with At-column dilution approach for peak transfer and the development of conditions for maximum peak trapping (trapping column chemistries and dilution solvents) are discussed.
The materials used in this work:
All solutions including mobile phase, standard, and test sample solutions were prepared as per related compounds procedure described in the USP Monograph for Lidocaine and Prilocaine Cream.2
ACQUITY UPLC Multi-dimensional System for Loop and Trap Interface with At-column dilution
|
1D system: |
ACQUITY UPLC H-Class Quaternary Solvent Manager (QSM) with Sample Manager Flow-Through-Needle (SM-FTN) and PDA Detector |
|
Dilution pump: |
ACQUITY UPLC Binary Solvent Manager (BSM) |
|
Column manager (CM-A): |
2D Column Tubing Kit (P/N 205000764) |
|
Loop size: |
1 mL (P/N 430002585) |
|
2D system: |
ACQUITY UPLC I-Class Binary Solvent Manager (BSM) with PDA and ACQUITY QDa Mass Detectors |
|
Mixer: |
50 μL (P/N 70000534) |
|
Column: |
SunFire, C18, 3.5 μm, 4.6 x 100 mm (P/N 186002553) |
|
Column temp.: |
40 °C |
|
Flow rate: |
1.5 mL/min |
|
Solvents A & B: |
Solution A and solution B as directed in the USP monograph for lidocaine and prilocaine cream2 |
|
Injection volume: |
50.0 μL |
|
Purge wash/sample wash: |
50:50 water/acetonitrile |
|
Seal wash: 90: |
10 water/acetonitrile |
|
PDA: |
210–400 nm, derived at 232 nm |
|
Time Flow (min) |
Flow (mL/min) |
%A |
%B |
%C |
%D |
Curve |
|---|---|---|---|---|---|---|
|
Initial |
1.500 |
67.0 |
33.0 |
0.0 |
0.0 |
Initial |
|
11.00 |
1.500 |
67.0 |
33.0 |
0.0 |
0.0 |
6 |
|
22.00 |
1.500 |
100.0 |
0.0 |
0.0 |
0.0 |
6 |
|
32.00 |
1.500 |
100.0 |
0.0 |
0.0 |
0.0 |
6 |
Additional three minutes for equilibration to the initial conditions.
|
Trap column: |
Oasis HLB, Direct Connect HP, 20 μm, 2.1 x 30 mm (P/N 186005231) |
|
Dilution solvent: |
2% ammonium hydroxide in water (Lines A and B of the dilution pump) |
|
Flow rate: |
2.0 mL/min |
|
At-column dilution: |
10:1 dilution ratio start at 2.5 min (Line A and B are set to 10% and 90%. This corresponds to 0.2 mL/min Line A for sample loading and 1.8 mL/min of Line B for dilution) |
|
Time (min) |
Flow (mL/min) |
%A |
%B |
Curve |
|---|---|---|---|---|
|
Initial |
0.200 |
50.0 |
50.0 |
Initial |
|
2.50 |
2.000 |
10.0 |
90.0 |
11 |
|
10.00 |
0.200 |
50.0 |
50.0 |
11 |
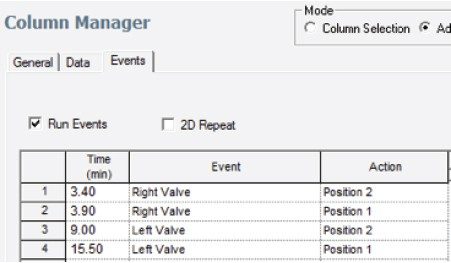
Timed events for heart-cutting impurity peak and transfer from loop/trap interface to 2D.
Initial valve positions:
Right valve: Position 1
Left valve: Position 2
|
Column: |
ACQUITY UPLC BEH C18 1.7 μm, 2.1 x 50 mm (P/N 186002350) |
|
Column temp.: |
40 °C |
|
Flow rate: |
0.5 mL/min |
|
Solvent A: |
0.5% Formic acid in water |
|
Solvent B: |
0.5% Formic acid in acetonitrile |
|
PDA: |
210–400 nm |
|
Time (min) |
Flow (mL/min) |
%A |
%B |
Curve |
|---|---|---|---|---|
|
Initial |
0.500 |
95.0 |
5.0 |
Initial |
|
9.00 |
0.500 |
95.0 |
5.0 |
6 |
|
14.00 |
0.500 |
5.0 |
95.0 |
6 |
|
15.00 |
0.500 |
5.0 |
95.0 |
6 |
|
15.50 |
0.500 |
95.0 |
5.0 |
6 |
|
Mass detector: |
ACQUITY QDa (Performance option) |
|
Ionization mode: |
ESI+, ESIMS |
|
acquisition time: |
9–14 min |
|
MS acquisition range: |
50–300 Da |
|
Sampling rate: |
10 pts/sec |
|
Capillary voltage: |
Pos: 0.8 kV, Neg: 0.8 kV |
|
Cone voltage: |
15 V |
|
Probe temp.: |
600 °C |
|
Data: |
Centroid |
Empower 3 FR4 CDS
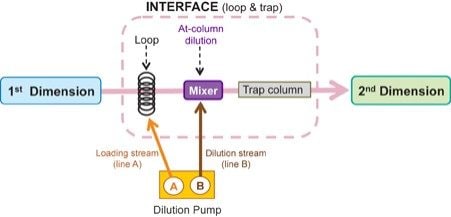
The approach in this study uses the heart-cutting technique with At-column dilution to transfer an impurity peak eluted in a non-MS compatible method run in the first dimension system, ACQUITY UPLC H-Class System, with UV detection. With a loop and trap configuration, a heart-cut volume is transferred to the holding loop, loaded onto a mixer, and diluted with an aqueous solvent. The peak is then refocused onto the trap column and transferred to the second dimension separation (Figure 1).
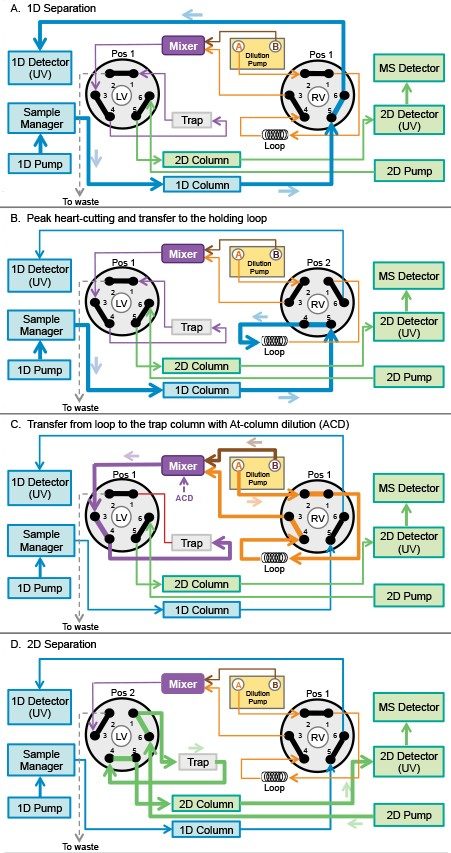
The workflow for fraction collection and transfer time is managed by coordinating the position of the two valves incorporated in the ACQUITY Column Manager and require specific configuration of fluidics connections (Figure 2). Each valve has two positions and six ports, each connected to different components of the multi-dimensional system. With both valves at position 1 (Figure 2A), flows from the 1D, dilution, and 2D pumps are independent of each other. In this case, the 1D separation is facilitated by the 1D pump and the dilution pump flows to the loop/trap interface and to waste. At the same time, the 2D pump flows through the 2D column and 2D detectors. When the right valve switches to position 2, the peak of interest is heart-cut from 1D analysis and transferred to the holding loop (Figure 2B). Meanwhile, the dilution pump moves to the dilution mode. Next, the heart-cut sample fraction is transferred from the holding loop to the trap column by switching the right valve back to position 1 (Figure 2C). In this step, the loading stream A of the dilution pump pushes the sample out of the loop to the mixer. At the same time, the dilution stream B dilutes the sample in the mixer with an aqueous solvent. This technique is known as At-column dilution (ACD). The ACD reduces composition of the organic solvent, focusing the analyte onto the trap column and producing higher retention, maximizing peak trapping.3,4 Finally, the MS compatible mobile phase from the 2D pump flows through the trap, transferring the trapped peak to the 2D UPLC column for analysis mass detection (Figure 2D).
The events for heart-cutting a peak must be synchronized and are programmed in the instrument method of the Empower Software as shown in the method conditions section.
USP monograph for lidocaine and prilocaine cream recommends using a Phenomenex Luna C18 with 4.6 x 100 mm, 3 µm column.2 We selected a Waters column with equivalent chemistry using a Waters Reversed-Phase Column Selectivity Chart5 and chose a SunFire C18 Column for analysis in 1D and an ACQUITY UPLC BEH C18 Column for UPLC separation in 2D, respectively.
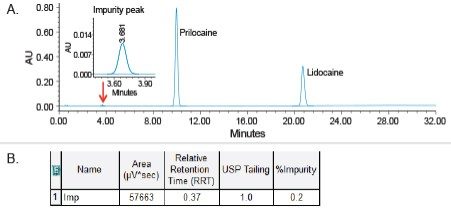
The USP related compounds method with phosphate buffer mobile phase was run with UV detection as a single-dimension method on an ACQUITY UPLC multi-dimensional system. The data showed presence of an impurity peak in the cream test preparation solution with retention time of 3.681 minutes (Figure 3A) and RRT of 0.37 relative to the prilocaine peak (Figure B). This peak was heart-cut from 1D and transferred to 2D for analysis by an ACQUITY QDa Mass Detector using the workflow described in Figure 2.
The impurity peak was heart-cut between 3.4 and 3.9 minutes, which equates to 0.75 mL volume for 1.5 mL/min flow rate in 1D. In this case, a 1 mL loop was chosen for holding the heart-cut peak volume before the dilution with an aqueous solvent. The dilution ratio was calculated based on the flow rate of the sample loading and dilution streams. The dilution ratio is crucial and if not set correctly can lead to peak distortion.3,4 With 10:1 ratio, sample was loaded at 0.2 mL/min and diluted at 1.8 mL/min, respectively. For a 1 mL loop and 0.2 mL/min loading stream, five minutes was required to ensure the peak was completely transferred from the loop to the trap column. Higher dilution ratios will reduce organic composition further, but will require longer transfer times.
Conditions for maximum peak trapping were optimized according to the method optimization protocol described in the Waters application note.4 The protocol was designed to evaluate the effect of dilution solvent (loading) pH, trap column chemistry, and mobile phase for 2D elution on the efficiency of peak trapping. The method optimization parameters and conditions are summarized in Figure 4.
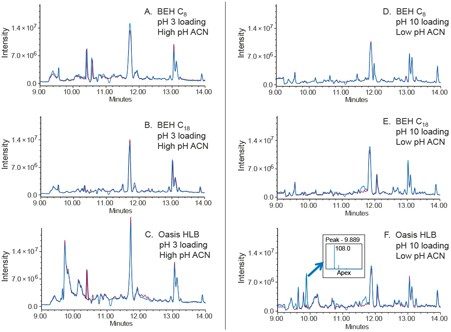
The heart-cut peak was transferred to 2D for analysis by mass detection using conditions described in Figure 4. The MS total ion chromatogram (TIC) data was collected for the diluent and the cream sample to differentiate background and system ions from the heart-cut peak. Example of the method optimization data for maximum peak trapping conditions processed using MS TIC in ESI+ mode is shown in Figure 5. The cream sample solution (blue trace) is overlaid with a diluent blank (red trace) to ensure that peaks found in the cream sample are not system impurities. Results from the method optimization demonstrates that the Oasis HLB Column with pH 10 dilution solvent and low pH with acetonitrile mobile phase provided the best trapping conditions for the impurity peak (Figure 5F). The mass spectrum data indicates that the peak of interest has a mass of m/z 108.0. This mass is a positively charged form [M+H]+ of the related compound of prilocaine HCl API known as o-Toluidine2 with a monoisotopic mass of 107.1 Da. Overall, the heart-cut peak has affinity for the most hydrophobic trap column (Oasis HLB), which indicates it is a hydrophobic compound.
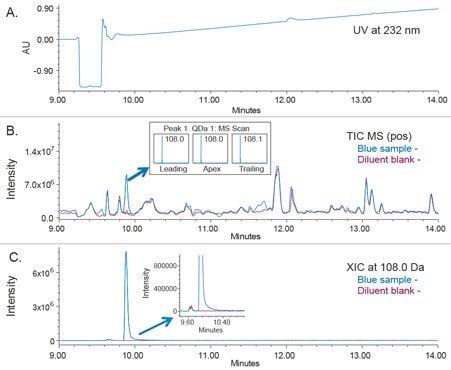
The 2D analysis of the o-Toluidine by UV and mass detection using an ACQUITY QDa Mass Detector is shown in Figure 6. The extracted ion chromatogram (XIC) at m/z 108.0 shows presence of the o-Toludine peak in the cream sample but not in the blank diluent (Figure 6C). This confirms that the peak observed in the cream assay preparation solution is not a system impurity. Moreover, the mass spectral data at the leading, apex, and tailing regions of the peak indicates presence of one mass with m/z 108.0. This demonstrates that o-Toludine is not coeluting with other peaks.
A multi-dimensional liquid chromatography, heart-cutting method was developed to couple mass spectrometry with a non-MS compatible USP method. Using this technology, an impurity peak observed in the cream assay preparation solution was successfully heart-cut from the 1D separation and transferred to the 2D under MS-compatible conditions to confirm identity by mass detection.
Many of the USP and registered methods utilize non-volatile buffers for assay testing during development or routine analysis of the pharmaceutical products. Any modifications to these methods will require a revalidation, which can be time consuming and costly. Utilizing the approach described within, these unmodified USP methods and any validated LC methods with non-volatile buffers can be directly coupled with mass spectrometry using heart-cutting technology. This technology will enable quick identification of unknown components by mass detection, eliminating the need to use additional instrumentation to isolate compound of interest, purify, and re-test under MS compatible conditions to obtain mass spectra data.
720006175, February 2018