In this application note, we outline a method utilizing the Waters ACQUITY UPLC System with Oligonucleotide Separation Technology (OST) Columns for the simultaneous annealing and purification of RNAi duplexes in a single step.
RNA interference (RNAi) is emerging as a new class of biopharmaceutical therapeutics for temporarily silencing genes and preventing protein translation. siRNA is a double stranded version of RNAi, which binds to RNA-inducing silencing complex (RISC). After cleavage of sense RNAi, part the RISC complex is activated, binds to a specific mRNA target, and, by cleaving, it interferes with protein production. This method of gene silencing is currently being utilized in a variety of animal studies and is receiving increased attention as a potential therapeutic strategy for humans.
A main challenge in developing therapeutics for humans remains the assurance of RNAi purity. The presence of certain related impurities may lead to unwanted, and potentially detrimental, off-target gene silencing. Major sources of impurities in siRNA duplexes originate from the complementary RNA strands’ synthesis (failure synthesis by-products).
Duplex RNAi is prepared from complementary single-stranded RNA (ssRNA) sequences. Both single RNA strands typically require purification prior to hybridization and annealing. Annealing should be performed using equimolar amounts of RNA, since the excess of non-hybridized ssRNA in the target duplex is undesirable and often associated with a decrease in siRNA therapeutic potency.
In this application note, we outline a method utilizing the Waters ACQUITY UPLC System with Oligonucleotide Separation Technology (OST) Columns for the simultaneous annealing and purification of RNAi duplexes in a single step. This method allows for sequential injection of complementary ssRNA molecules, which tightly focus on the column, anneal, and elute as a duplex.
The duplex can be collected by appropriate heart-cutting of the main peak, which yields a highly pure and stoichiometric duplex in a single step, as well as dramatically reduces the time and reagents needed to prepare the duplexes. Reduction in time and reagents coupled with the high purity of our method significantly lowers siRNA purification costs and increases production throughput.
RNAi complementary strands (5' – UCG UCA AGC GAU UAC AAG GTT – 3' and 5' – CCU UGU AAU CGC UUG ACG ATT – 3') were purchased from Integrated DNA Technologies and reconstituted in 0.1 M triethylammonium acetate (TEAA), which was purchased as a 2 M solution from Fluka and diluted in 18 MW water to yield concentrations of approximately 2 nmol/μL.
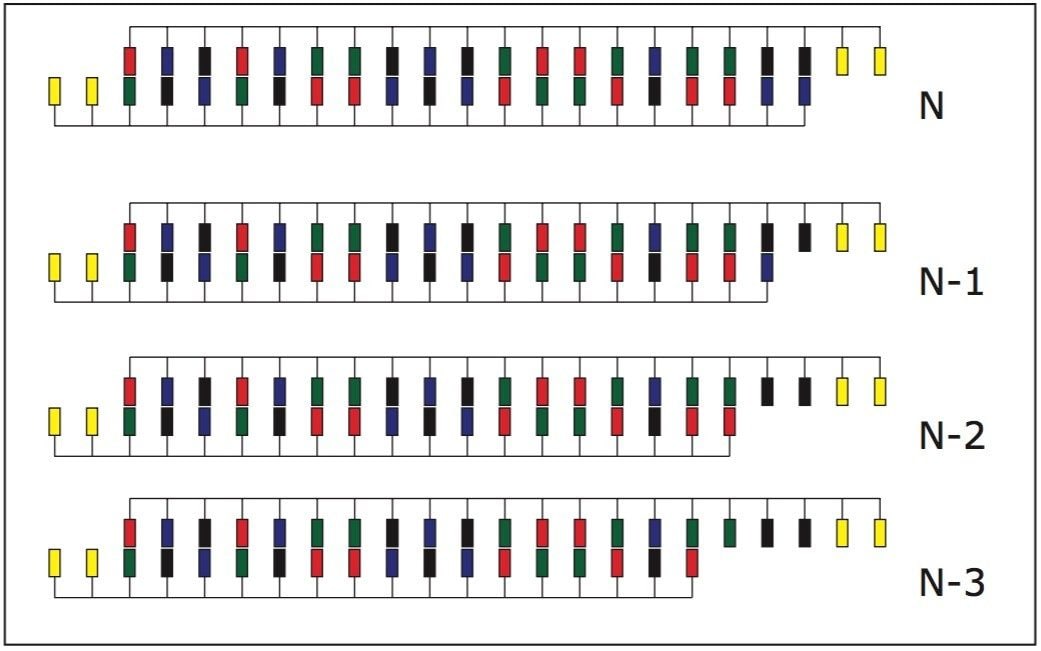
An aliquot of one set of complementary strands was purified as ssRNA1 and one strand was partially digested with the exonuclease phosphodiesterase II to generate a 5' truncated ladder or RNAi.
We determined the ability of UPLC to resolve the desirable siRNA duplex from its truncated forms (that were formed due to presence of failure synthetic RNA strands in the annealing mixture). In order to demonstrate UPLC’s resolving performance for RNA duplexes, we utilized purified upper RNAi strand and partially digested lower strand. Upon annealing, a ladder of siRNA duplexes was formed with partially 5' truncated lower RNAi strands, as shown in Scheme 1.
|
LC system: |
Waters ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC OST C18, 2.1 x 50 mm, 1.7 μm (P/N 186003949) |
|
Column temp.: |
20 °C |
|
Flow rate: |
0.2 mL/min |
|
Mobile phase A: |
25 mM HAA, pH 7.0 |
|
Mobile phase B: |
100% Acetonitrile |
|
Gradient: |
30% to 40% B in 10.0 min (1% ACN/min) |
|
Detection: |
PDA Detector, 260 nm SQ, 600 to 2000 Da |
|
MS system: |
Waters SQ Detector |
|
Mode: |
ES- |
|
Capillary: |
3.0 kV |
|
Cone: |
28.0 V |
|
Extractor: |
3.0 V |
|
RF: |
0.1 V |
|
Source temp.: |
150 °C |
|
Desolvation temp.: |
350 °C |
|
Cone gas flow: |
31 L/h |
|
Desolvation gas flow: |
700 L/h |
Figure 1 shows the chromatographic results from the ACQUITY UPLC System and OST Column technology, which successfully resolved truncated siRNA duplexes from full-length duplex and single-stranded RNAi species. The separation was performed at 20 °C to maintain siRNA in a duplex form.
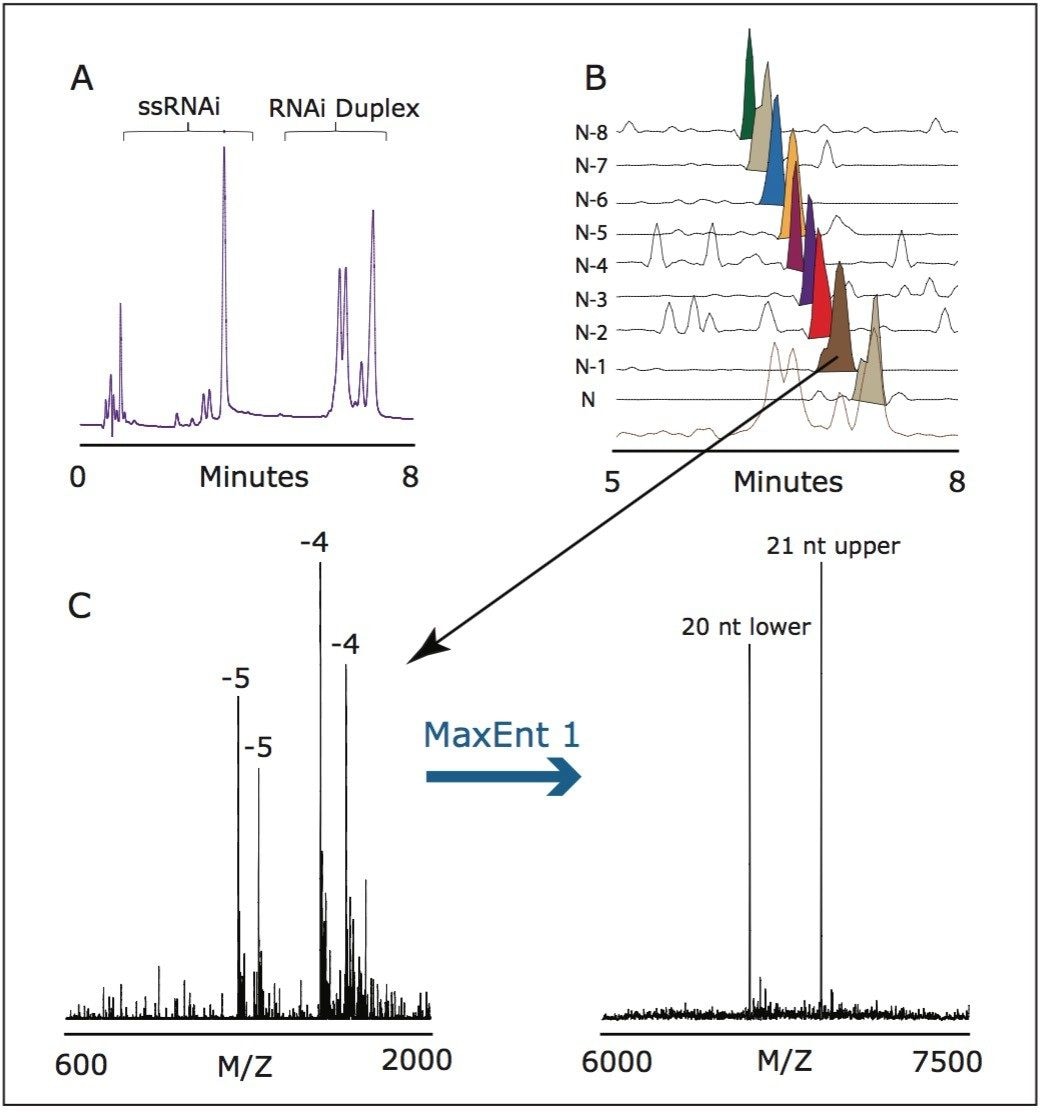
Figure 1. UPLC analysis of RNAi duplex mixture.
Panel A: UPLC PDA Detector 260 trace of full-length upper strand and truncated lower strand.
Panel B: SQ Detector TIC and SIC for UPLC analysis of RNAi duplexes.
Panel C: Representative MS spectrum for RNAi duplex with MaxEnt1 deconvolution.
Using MS-compatible mobile phase comprised of hexylammonium acetate, we identified each eluting duplex peak by the corresponding mass of complementary RNAi strands and confirmed the elution order of the impurities. Extracted selected ion chromatograms, shown in Figure 1, indicate that retention time correlates with the length of the truncated complementary strand. The full length siRNA duplex eluted after the partially truncated duplexes.
Based on the ACQUITY UPLC System’s ability to provide impressive resolution of RNAi duplexes, we determined its utility for semi-preparative purification of siRNA duplexes as prepared from crude mixtures of ssRNA.
To accomplish this, we first mixed complementary ssRNA stoichiometrically and annealed the resulting mixture by heating the sample to 90 °C followed by cooling slowly to room temperature. This mixture was then separated via HPLC on an analytical-scale column and the appropriate duplex fraction was collected, as shown in Figure 2.
|
LC system: |
Waters Alliance Bio HPLC System |
|
Column: |
Waters XBridge OST BEH C18, 4.6 x 50 mm, 2.5 μm |
|
Column temp.: |
20 °C |
|
Flow rate: |
1.0 mL/min |
|
Mobile phase A: |
0.1 M TEAA, pH 7.0 |
|
Mobile phase B: |
20% ACN in A |
|
Gradient: |
25% to 75% B in 30 min |
|
Detection: |
PDA, 260 nm |
We found quantitative conversion to the desired siRNA duplex with good resolution of the main product from both single-stranded and duplex impurities. Analysis of the collected fraction was obtained using UPLC with the PDA and SQ Detectors; MS detection indicated 98% purity of collected siRNA. No single-stranded contaminants were detected by UPLC analysis (data not shown).
To further evaluate the utility of our method, we investigated the possibility of on-column annealing of crude complementary ssRNA. To accomplish this, we prepared solutions of each crude complementary strand in 0.1 M TEAA and verified the concentration by injection of a small amount of each solution.
Since each strand is of the same length, with similar extinction coefficients, the use of integrated peak area was found to be sufficient for calculating the desirable injection volumes to introduce an approximately stoichiometric amount of RNA on column.
The first RNA strand was injected onto the column under initial gradient conditions. Immediately after, the second complementary strand was injected and the gradient elution was initiated.
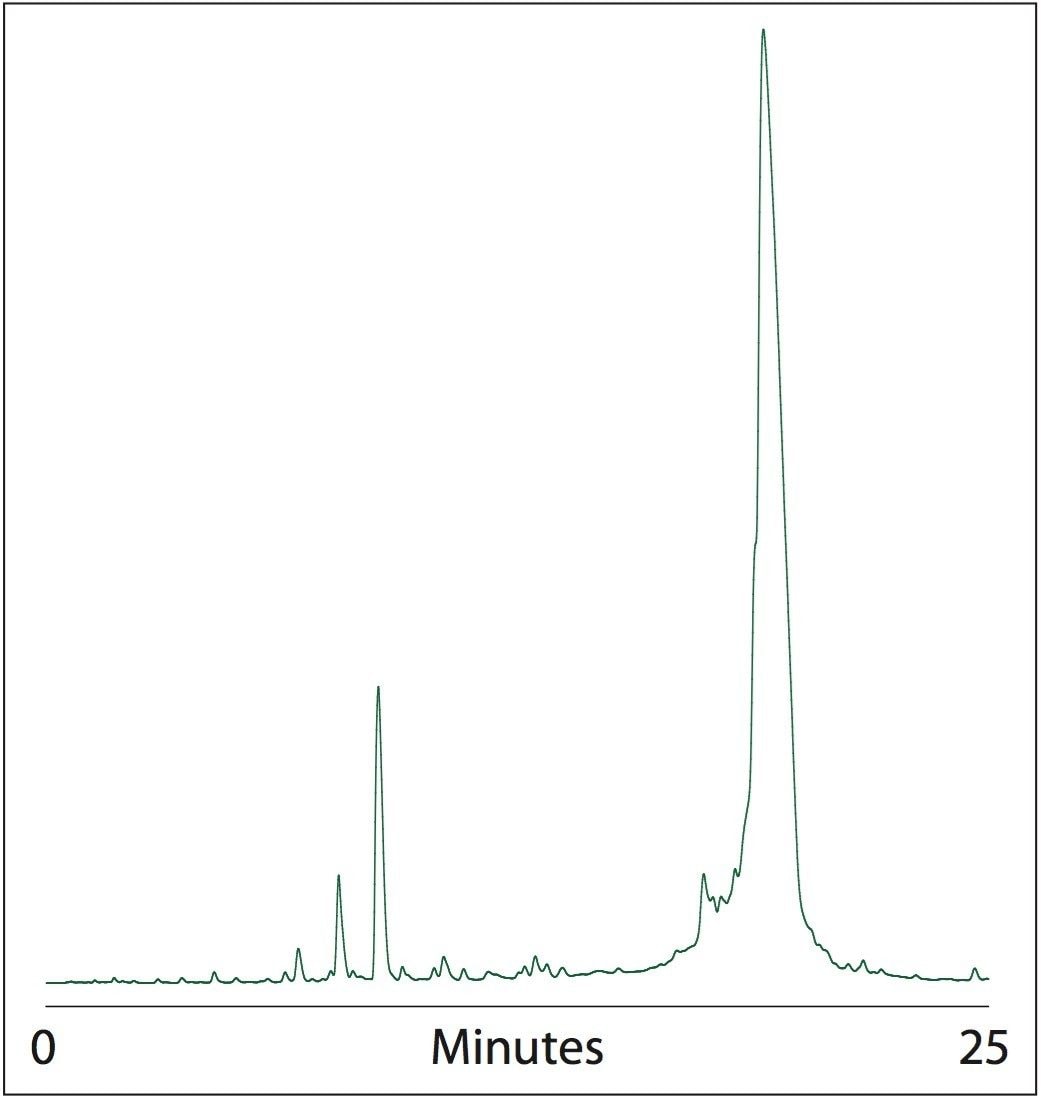
The chromatogram in Figure 3 reveals that both strands anneal nearly quantitatively on-column and are eluted as duplex siRNA. This is probably due to tight spatial focusing of complementary strands on the head of the column. Small excess of one single-stranded RNAi eluted prior to the main peak, as expected. Truncated RNA duplexes were resolved from the target siRNA.
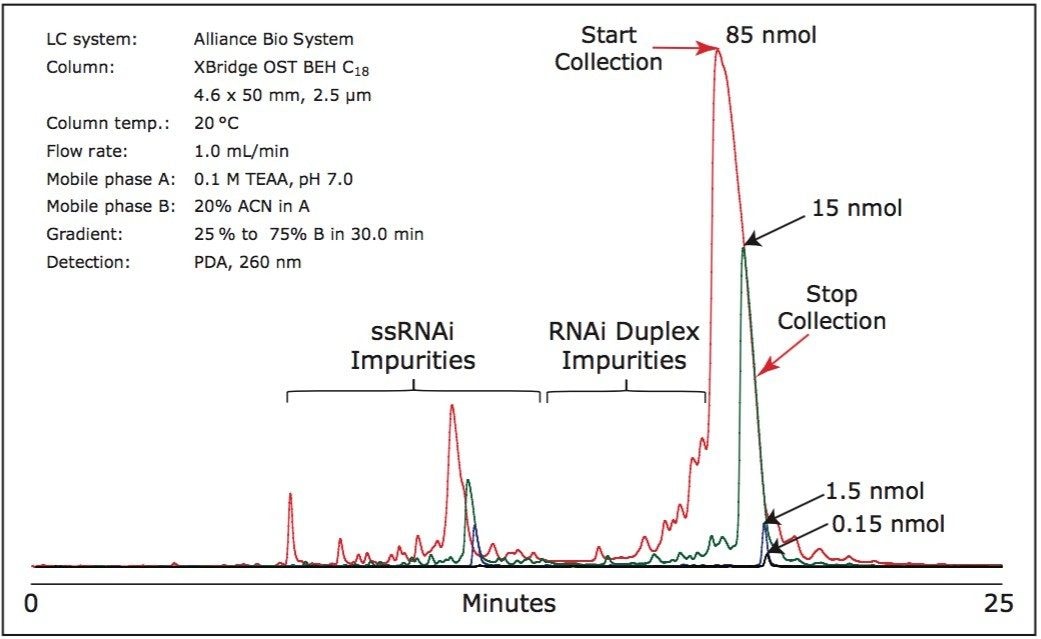
Figure 3 illustrates the siRNA purification with on-column annealing on three different mass loads. The method for siRNA purification was scaled up to ~85 nmol using analytical column.
The on-column annealing siRNA purification methods represent a significant improvement over the earlier presented RNAi purification.2 By eliminating the need for separate purification steps for each complementary strand, and annealing the strands on-column, our method allows the researcher to substantially reduce the time needed for sample preparation.
Following purification, collected fractions were analyzed with UPLC analysis (Figure 4), which was done in the highly-resolving mobile phase HAA at 20 °C to preserve the duplex. We also analyzed the duplex at 60 °C, which fully denatured the duplex and generated two single-stranded counterparts. Both analysis techniques indicated that the fraction collected contains the desired duplex and that the purity is greater than 98%.
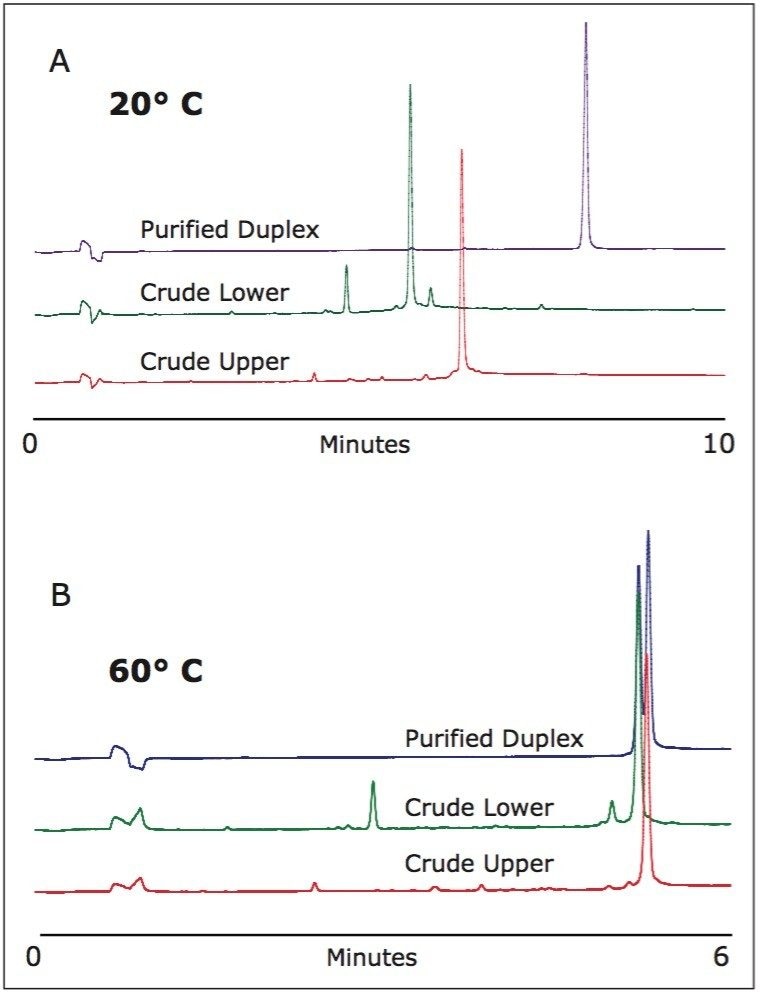
|
LC system: |
ACQUITY UPLC System |
|
Column: |
ACQUITY UPLC OST C18, 2.1 x 50 mm, 1.7 μm |
|
Column temp.: |
20 °C |
|
Flow rate: |
0.2 mL/min |
|
Mobile phase A: |
25 mM HAA, pH 7.0 |
|
Mobile phase B: |
100% Acetonitrile |
|
Gradient: |
30% to 40% B in 10.0 min, (1% ACN/min) |
|
Detection: |
PDA, 260 nm |
In this application note, we have described a novel method for analysis and purification of double stranded siRNA. Using the Alliance Bio HPLC and ACQUITY UPLC Systems with ACQUITY UPLC and XBridge OST Columns, we were able to efficiently resolve full-length siRNA duplexes from shorter truncated duplexes. The method can easily be scaled from analytical to preparative, allowing for fast purification of siRNA prior to gene silencing experiments.
The method utilized non-denaturing, mass spectrometry-compatible mobile phase comprising hexylammonium acetate and acetonitrile. The separation of single-stranded RNAi impurities and siRNA truncated duplexes was monitored by UV and MS. The retention order of impurities was confirmed by MS data. Non-denaturing mobile phases and low separation temperatures are necessary to maintain the stability of non-covalent complexes (duplex RNA) throughout the analysis.
This application note proposes a novel approach for purification of duplex siRNA using on-column annealing of RNA strands rather than purification of RNAi in single-stranded form (followed by off-line annealing). The presented method allows for high yields and purity of the desired duplex in a dramatically shorter time period. Volatile mobile phases allow for easy removal of mobile phase without the need for additional desalting.
The proposed purification strategy has potential to significantly improve the productivity of siRNA manufacturing. It allows manufacturers to ship the custom made siRNA product within a single day, which is often not attainable with the traditional purification strategies. Faster manufacturing of high quality siRNA probes will help to facilitate the adoption of silencing RNA technology.
720002800, October 2008