
Ion mobility data independent mode of acquisition (HDMSE) provides a mechanism to clean up and resolve metabolite data. HDMSE approaches provide fast, high quality datasets in a routine and efficient manner. This application note outlines and focuses on the benefits and the ease of use of ion mobility for the application of metabolite identification.
Drug Metabolite identification relies on confident detection and structural characterization of metabolites. Obtaining high quality data in the presence of diverse and complex matrices (tissue/fluid type/species) can be challenging. It is essential to track and identify these metabolites, especially given the existence of biological mechanisms by which two or more isobaric metabolites can be formed. By measuring CCS accurately it is possible to track these metabolites and glean insight into how the biotransformation has changed the molecular shape. Increasingly, scientists are invoking advanced analytical tools such as ion mobility to help solve these challenges.
Historically, ion mobility-equipped systems required complex and separate drift time enabled software packages for processing and data interpretation. UNIFI Software and the Vion IMS QTof Mass Spectrometer now implement this in a straightforward and user friendly manner, enabling easy access to cleaner data and accurate CCS measurements.
The benefits of ion mobility include: generation of the highest quality data possible with cleaner spectra; access to CCS values for all ions; and the ability to go beyond mass resolution, in order to obtain additional information about metabolite isomers.
This application note outlines and focuses on the benefits and the ease of use of ion mobility for the application of metabolite identification. Here we highlight a data independent ion mobility data collection approach (HDMSE), to track and resolve, including tracking metabolites across different chromatographic conditions. We include examples of chromatographic co-elution which would otherwise have resulted in mixed product ion spectra.
Nefazodone and buspirone (Figure 1) were incubated (10 µM) with cryopreserved rat hepatocytes and relevant cofactors at 37 °C for 0, 15, 30, 60, 120, and 240 minutes. The incubations were terminated by addition of an equal volume of ice cold acetonitrile, centrifuged, and the supernatant was analyzed. To demonstrate the value of ion mobility separation in more complex matrices, supernatant was further diluted (1 in 10) in urine.
|
System: |
ACQUITY UPLC I-Class (FTN) |
|
Column: |
ACQUITY UPLC HSS T3 2.1 x 100 mm, 1.8 μm (P/N 186003539) |
|
Total run time: |
Samples analyzed with 5, 10, and 15 min chromatography |
|
Vials: |
Waters Maximum Recovery |
|
Column temp.: |
45 °C |
|
Sample temp.: |
6 °C |
|
Injection volume: |
0.1–10 μL |
|
Flow rate: |
0.6 mL/min |
|
Mobile phase A: |
Water + 0.1% formic acid |
|
Mobile phase B: |
Acetonitrile + 0.1% formic acid |
|
Time |
%A |
%B |
Curve |
|---|---|---|---|
|
0.0 |
98 |
2 |
– |
|
7.5 |
40 |
60 |
6 |
|
8.0 |
0 |
100 |
6 |
|
8.5 |
98 |
2 |
11 |
|
MS system: |
Vion IMS QTof |
|
Ionization mode: |
ESI+ |
|
Acquisition mode: |
HDMSE, 0.1 sec scan time |
|
Source temp.: |
120 °C |
|
Desolvation temp.: |
450 °C |
|
Desolvation gas: |
800 L/hr |
|
Capillary voltage: |
1.0 kV |
|
Cone voltage: |
40 V |
|
Reference mass: |
Leucine enkephalin [M+H]+ m/z 556.27658 |
Ion mobility mass spectrometry is often perceived as more complicated than traditional LC/MS workflows. Within the UNIFI Software and the Vion IMS QTof Mass Spectrometer, however, the process is fully integrated and CCS values are automatically calculated during method processing for all components detected within a sample.1 CCS measurements are calibrated as part of a simplified instrument setup routine.
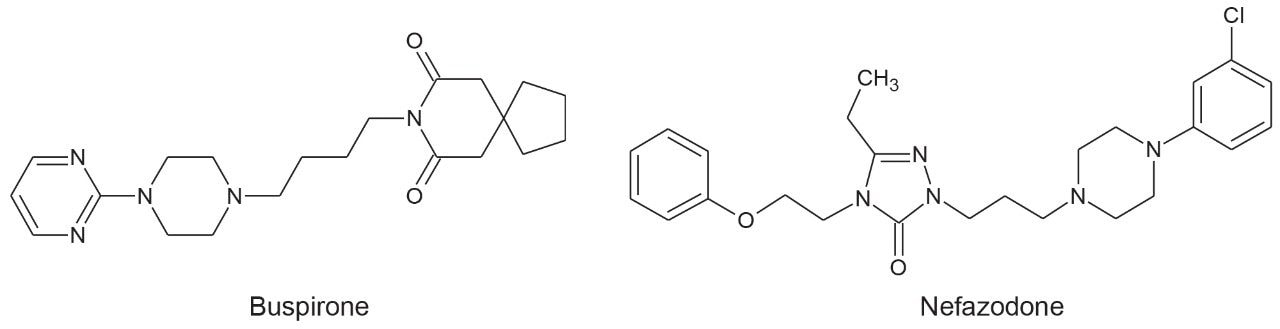
Ion mobility enables high spectral clarity. It does this through alignment of precursors and fragment ions on the basis of both retention time (tR) and drift time (dt), facilitating rapid interrogation of targeted precursors as well as fragment ions. Ion mobility-resolved spectra are particularly useful in complex samples. Shown in Figure 2 are the precursor and fragment ion (HDMSE) spectra for the dihydroxylated glucuronide metabolite of buspirone, with the protonated species identified at m/z 594.28. The spectra on the left (Figure 2A) is tR aligned and the spectra on the right (Figure 2B) is both tR and (ion mobility) dt aligned. The latter shows a significant clean up and a large number of background ions from the urine and hepatocyte matrix have been removed. More importantly, however, within the high energy spectrum the fragment ions relating to the dihydroxylated glucuronide metabolite are now visible and can subsequently be assigned during data processing.
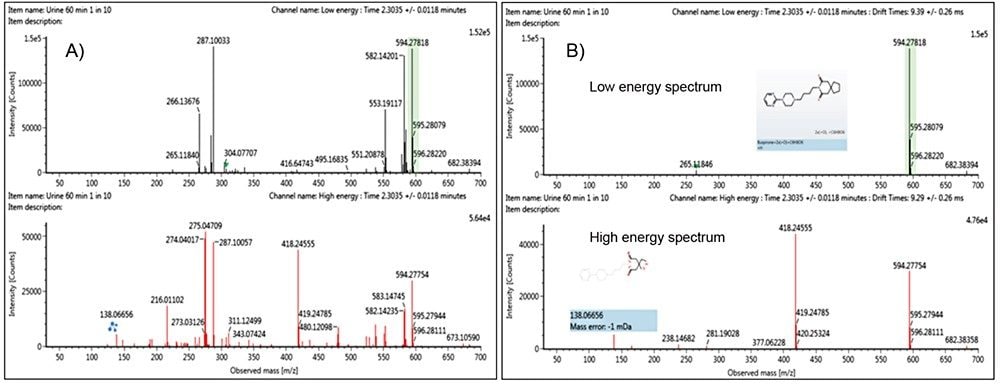
Ion mobility information can also be used to generate ion mobility-filtered extracted ion chromatograms (XICs) with greater clarity than those obtained from extraction of m/z values alone. This yields more selective XICs and improvements in signal to noise (S/N) ratios. These higher quality chromatograms of metabolites are important for confident identifications and achieving lower limits of detection. Shown in Figure 3 are XICs for the dihydroxylated glucuronide of nefazodone with (A) and without (B) the incorporation of ion mobility into the XIC selection. Highlighted on the XICs are the conditions under which the XIC were performed and the measured peak to peak S/N ratios. The S/N ratio increased by three fold when the XIC was performed with the addition of drift time selection compared to m/z alone. In addition, the sample was diluted further to 1 in 100 (with a 0.1 µL injection volume) mimicking in vivo sample sets where reaching lower levels of detection for low abundance metabolites can be challenging.
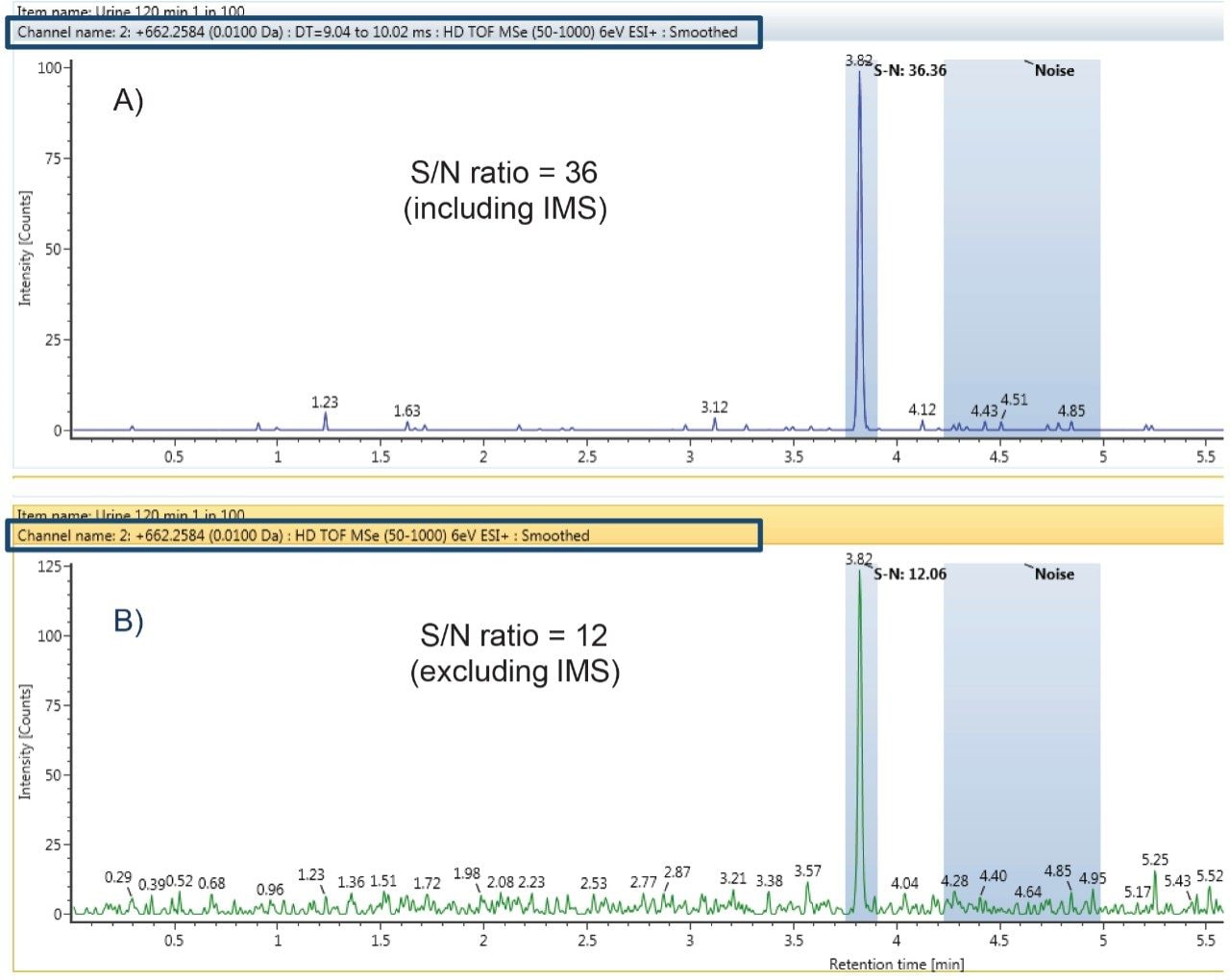
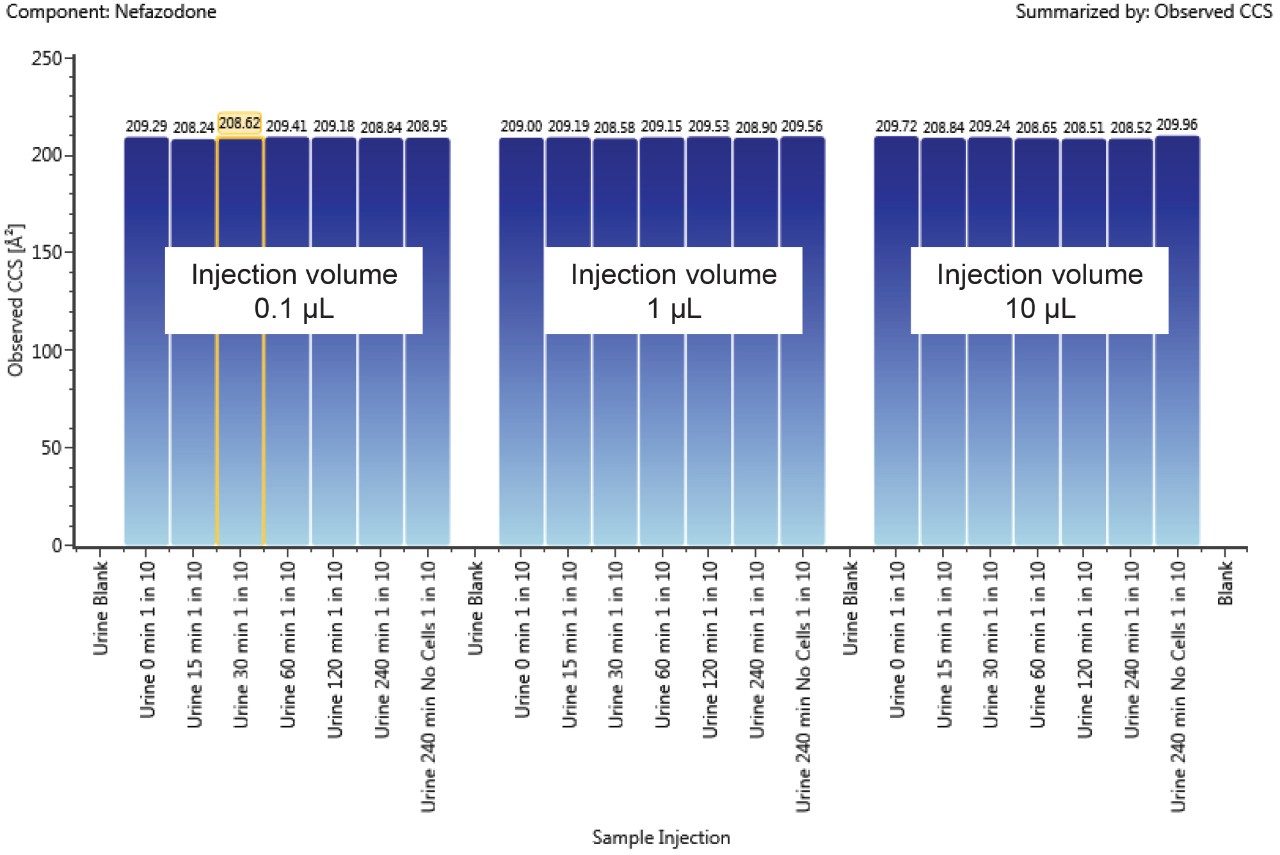
A component’s CCS value can be used to track isobaric metabolites across chromatographic conditions and to resolve closely or co-eluting metabolites. The CCS value is a reliable and consistent value, independent of matrix and concentration of the ion. Figure 4 shows the trend plot of CCS for nefazodone throughout the incubation experiment. Over the time course nefazodone varies significantly in concentration (over three orders) and is present in a complex matrix thus demonstrating the robustness of the CCS measurement as an identification point. The %RSD of the measurement over the 250 hour experiment was 0.2%.
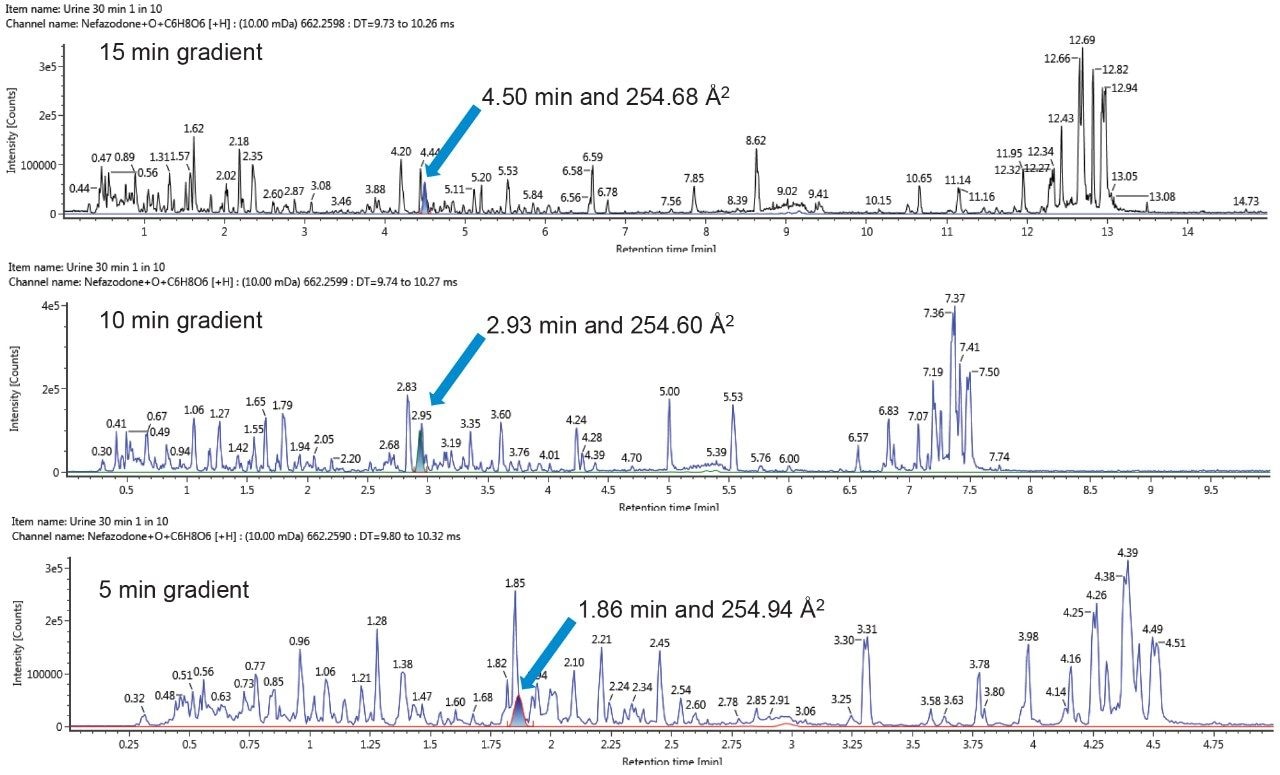
During the lifetime of a drug, a candidate molecule may be analyzed under several different LC-MS conditions as it progresses from discovery, to development, to preclinical trials and beyond. The characteristic CCS values of metabolites, together with fragmentation patterns, allow for them to be tracked and identified, thus easing the burden of metabolite identification under differing conditions.2 Figure 5 shows an example of tracking a metabolite, a hydroxylated glucuronide of nefazodone (259.65 Å), across different methods. Fast acquisitions are typically employed in early discovery experiments. When, however, it may be necessary to chromatographically resolve co-eluting metabolites or when analyzing radio-labelled sample sets a longer acquisition time would typically be utilized.
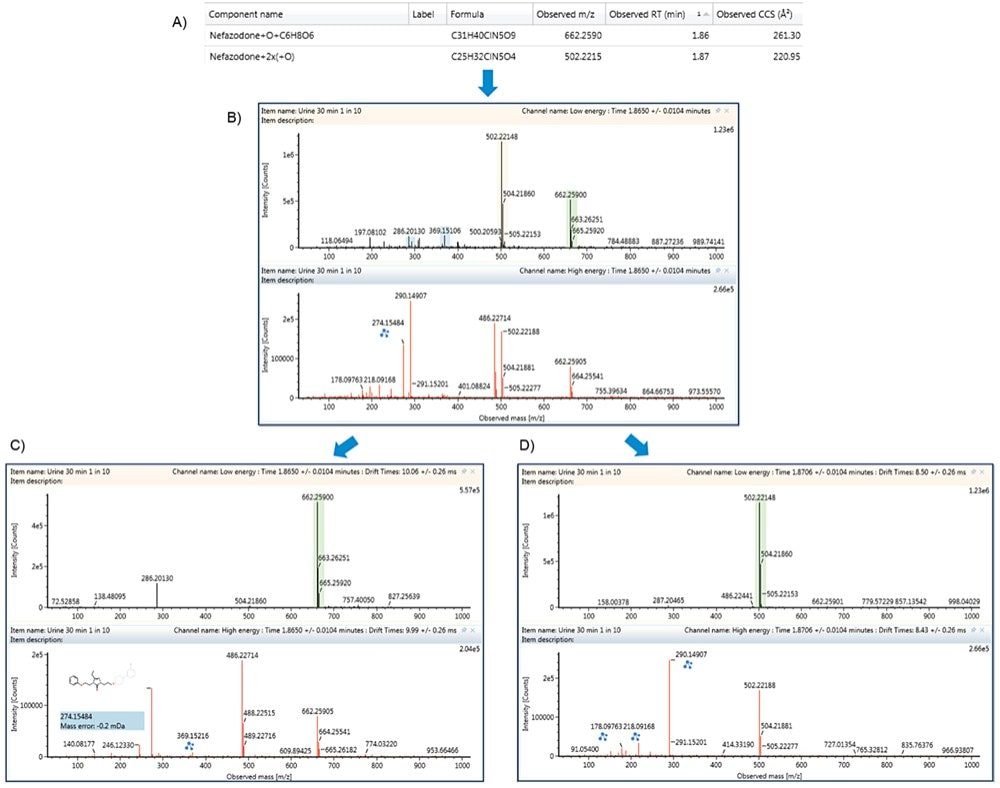
The challenges with data dependent acquisition (DDA) modes for multiple co-eluting peaks, especially at varying or low concentrations, is that metabolites may be missed, in that the quadrupole may not switch to perform MS/MS if intensities of ions are too low or if there are several analytes of interest closely eluting. HDMSE provides a straightforward way to both collect all fragment ions and resolve them into MS/MS quality spectra, without the concern for lost identifications. The dihydroxylated and hydroxylated glucuronides of nefazodone co-elute at 1.86/1.87 min giving a mixed precursor and fragment ion spectrum as shown in Figures 6A and 6B. The metabolites, however, have different CCS measurements of 220.95 Å2 and 261.30 Å2, respectively, which allows their corresponding fragment ion spectra to be discriminated by the software. This limits incorrect assignment of localization of the transformations. The resolved precursor and fragment ion spectra for the hydroxylated glucuronide and dihydroxylated metabolites of nefazodone are shown in Figures 6C and 6D, respectively.
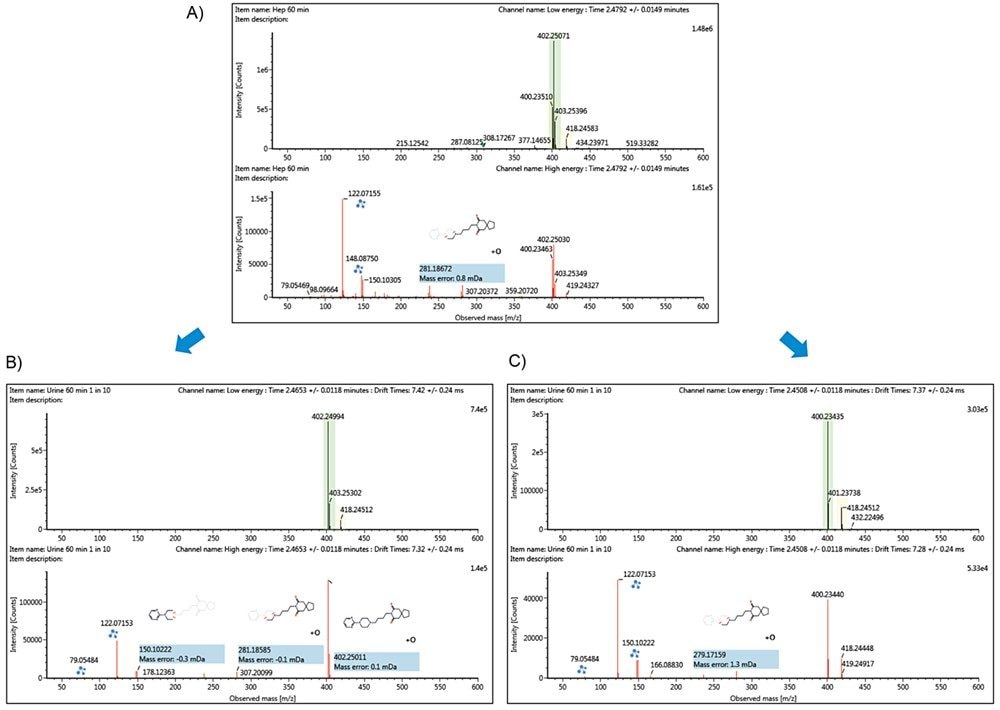
A similar example was observed with buspirone in which three metabolites corresponding to the dihydroxylated, hydroxylated and desaturated + hydroxylated metabolites at m/z 418.25, 402.25, and 400.24 respectively, co-elute. This results in a mixed high fragment ion spectrum (Figure 7A). The observed doublets at m/z 236.12/238.14 and 279.17/281.19 in the fragment ion spectrum suggests the presence of both the hydroxylated and desaturated + hydroxylated metabolites of buspirone. These spectra were deconvolved by dt alignment resulting in the clear and unambiguous spectra shown in Figure 7B and 7C for the hydroxylated and desaturated + hydroxylated metabolites of buspirone. On closer inspection, the doublet at m/z 148.09 and 150.10 is thought to be correct as observation of an additional hydroxylated metabolite of buspirone at a later retention time which undergoes no co-elution also forms this pair of fragment ions.
In addition, there is also a clear benefit to ion mobility separation for isobaric and overlapping metabolites. Previously described work by R. Clayton et al., on co-eluting nefazodone glucuronidative species has shown how spectra can be deconvolved and resolved aiding in the elucidation and assignment in the position of transformation metabolites.3 This work demonstrates the ability to go beyond mass resolution and obtaining additional information about metabolite isomers.
Ion mobility data independent mode of acquisition (HDMSE) provides a mechanism to clean up and resolve metabolite data. HDMSE approaches provide fast, high quality datasets in a routine and efficient manner. The examples shown have highlighted benefits including generating the highest quality data possible with cleaner spectra for 1) the removal of matrix interference, 2) co-elution of metabolites, and 3) tracking isobaric metabolites under different chromatographic conditions using the automatically generated CCS values.
Waters Metabolite Identification Application for UNIFI in combination with ion mobility separation redefines the way that users can interact with data collected in drug metabolism studies. By incorporating ion mobility in a routine manner it is possible to reduce labor and time intensive data interrogation, ultimately providing clarity and confidence to datasets and system productivity to laboratories.
720006121, October 2017