
This application note presents an efficient and cost effective UHPLC-MS method using the ACQUITY Arc UHPLC System coupled with the ACQUITY QDa Mass Detector to screen and quantify carbamates in raw agricultural commodities, drinking and surface water, and soil, as an alternative to post column derivatization used in the Chinese GB standard (NY/T-761 2008).
Using DisQuE dSPE cleanup with a “dilute and shoot” sample preparation procedure, all analytes were detected using the ACQUITY QDa Mass Detector. No derivatization or time-consuming sample preparation was required, thus allowing for the rapid quantitation of carbamate pesticides on a single detector.
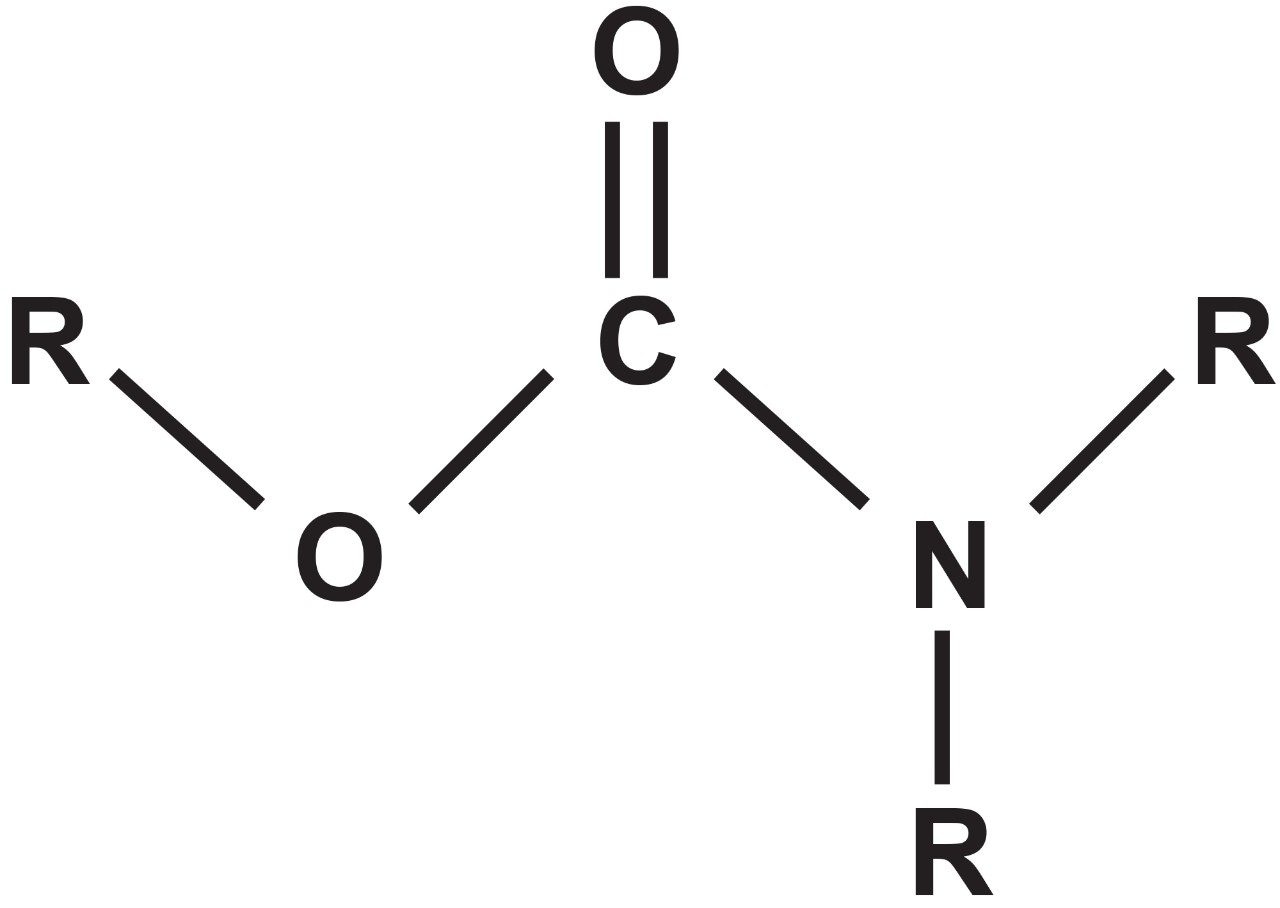
Carbamate pesticides are derived from carbamic acid and kill insects in a similar fashion as organophosphate insecticides. They are widely used in agricultural production and are thus transferred to food which necessitates a need for rapid analytical methods to screen and quantify carbamates in raw agricultural commodities, drinking and surface water, and soil. The general formula of the carbamates is shown in Figure 1, where “R” represents the alkyl or aryl groups.
Currently, the official Chinese GB standard (NY/T-761 2008) used for carbamate analysis requires HPLC with post-column derivatization with fluorescence detection.1 As routine testing requires the analysis of carbamate pesticides in a variety of complex food matrices, LC-MS/MS methodologies offer a confirmatory analysis following the AOAC standard 2007.012 or CEN standard method 15662:2008.3
Mass detection offers increased selectivity and sensitivity when compared to optical detectors such as ultraviolet (UV), photodiode array (PDA), fluorescence (FLR), refractive index (RI), or electrochemical detection for identifying target compounds in complex matrices. While mass spectrometry (MS), especially tandem quadrupole MS, has been used for trace detection of contaminants, the cost of MS/MS systems can be prohibitive for many routine analysis laboratories. Typical carbamates testing does not necessarily require the level of sensitivity offered by MS/MS systems to meet the regulatory residual limits of 10 μg/kg.
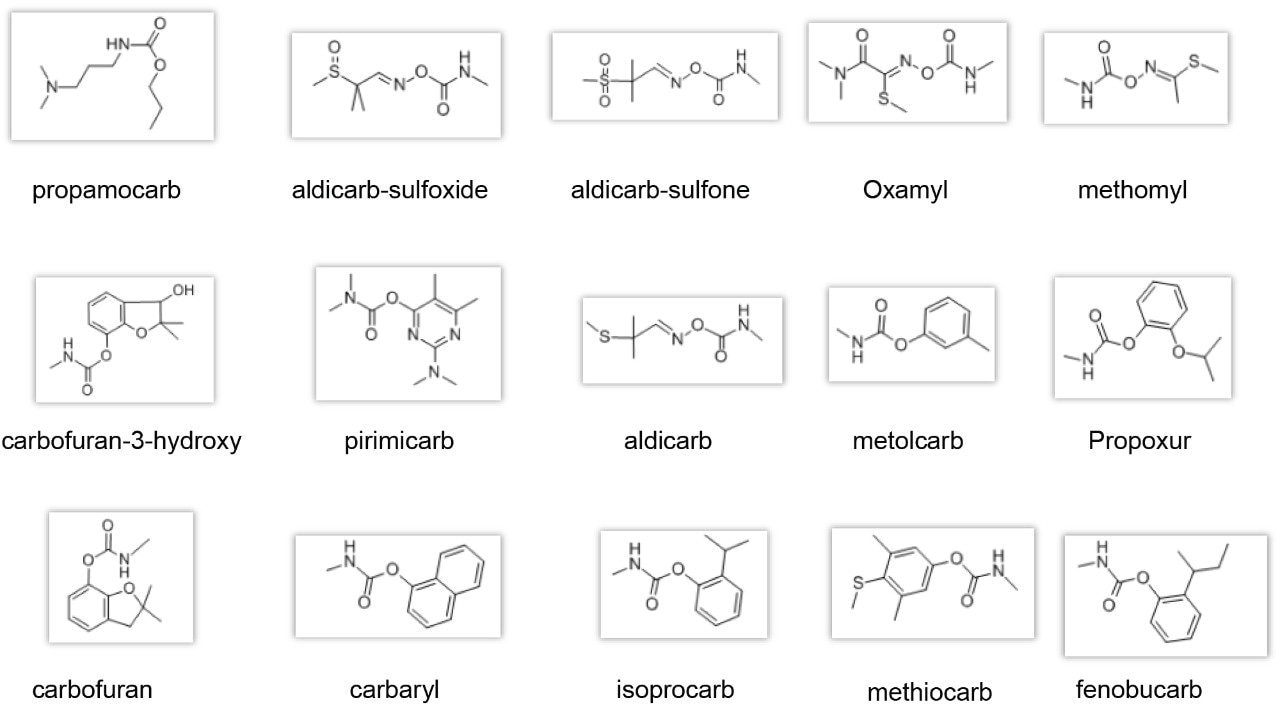
In this application note we show an efficient and cost effective UHPLC-MS method using Waters ACQUITY Arc UHPLC System coupled with the ACQUITY QDa Mass Detector for the quantification of carbamate pesticides, as an alternative to post column derivatization used in the Chinese GB standard (NY/T-761 2008).1 Figure 2 shows the structures of the targeted carbamates. The ACQUITY QDa Detector enables implementation of mass detection capabilities to an LC workflow that currently employs optical detectors. The selectivity of mass detection allows for low level detection of carbamates, enabling simpler sample preparation protocol for complex matrices when compared to HPLC-FLR method.
|
UHPLC system: |
ACQUITY Arc |
|
Column: |
CORTECS T3 2.7 μm, 3 x 150 mm |
|
Column temp.: |
40 °C |
|
Injection volume: |
5 μL |
|
Flow rate: |
1.0 mL/min |
|
Mobile phase A: |
Water with 0.1% formic acid, 2 mM ammonium formate |
|
Mobile phase B: |
Methanol with 0.1% formic acid, 2 mM ammonium formate |
|
Gradient: |
5% B initial and hold to 1.0 min, linear gradient to 90% B at 5.0 min, hold to 8.0 min, back to 5% B immediately, hold and re-equilibrate until 12 min. |
|
MS system: |
ACQUITY QDa |
|
Ionization mode: |
ESI+ |
|
Capillary voltage: |
0.5 kV |
|
Source temp.: |
120 °C |
|
Probe temp.: |
600 °C |
|
Sampling rate: |
10 Hz |
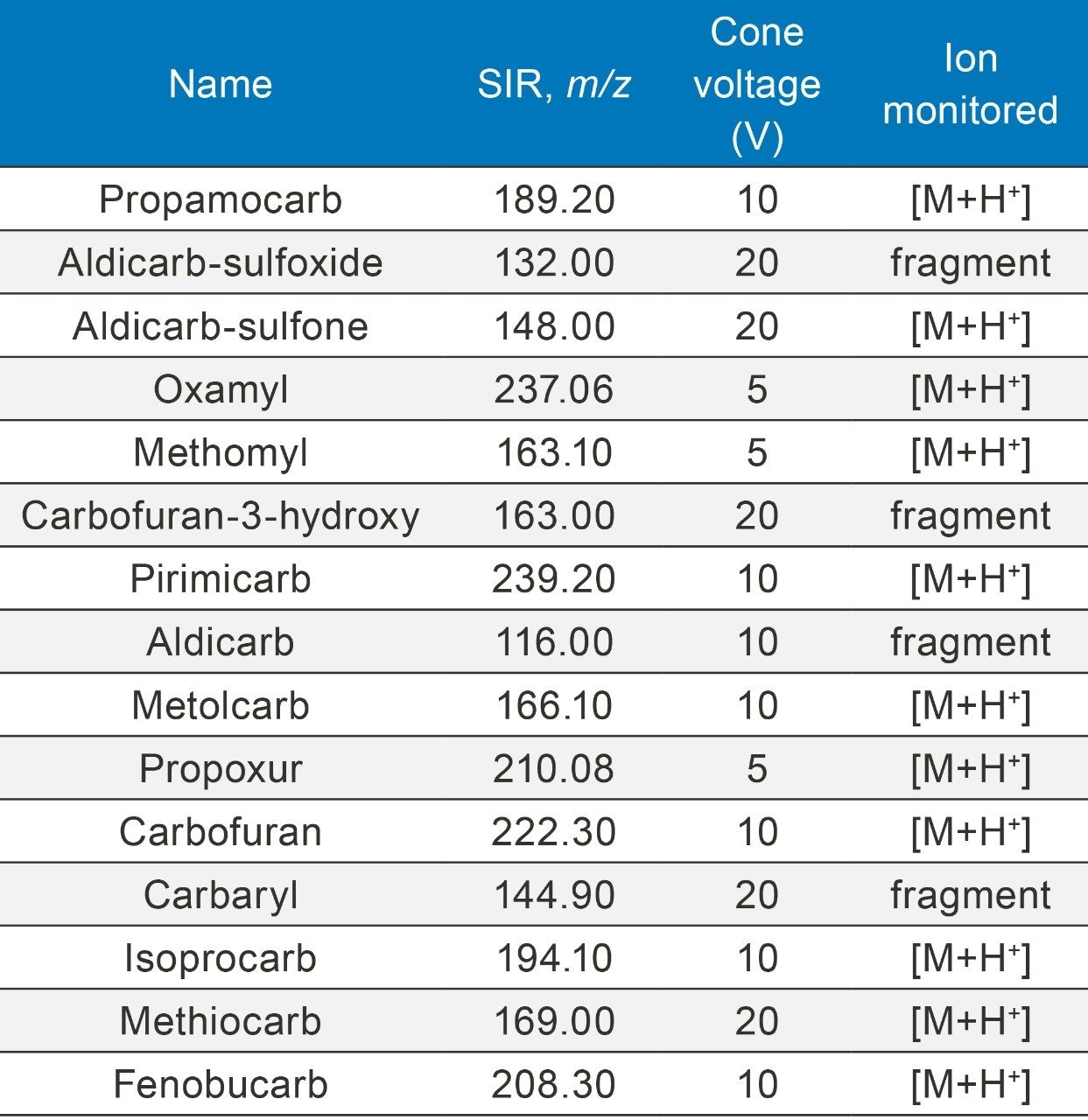
MS cone voltage and ion masses monitored in this study are presented in Table 1.
Sample extraction
Weigh 15 g of the sample into 50-mL tube, add 10 mL of water. Note that the amount of water added depends on the water content of the sample (e.g. it is not necessary to add any water to a sample when its water content is more than 80%). Add 15 mL of 1% acetic acid in acetonitrile (ACN), and shake for 1 min. Add contents of DisQuE Pouch (p/n 186006812) and shake vigorously for 1 min, then centrifuge at 3000 rcf for 10 min. Transfer 1 mL of the supernatant into the DisQuE QuEChERS 2-mL Tube (p/n 186008071) for cleanup. Vortex tube for 1 min and centrifuge again at 3000 rcf for 10 min. Dilute the supernatant four times with water, filter the diluents, and transfer to the vial for UHPLC-MS analysis using the ACQUITY Arc System and ACQUITY QDa Detector.
Preparation of standards
Matrix-matched calibration curves allow for accurate quantification of carbamates fortified in the matrix at regulatory limits. Calibration curves ranged from 1 to 100 μg/L for each target compound. Linearity for target analytes is shown in Table 2.
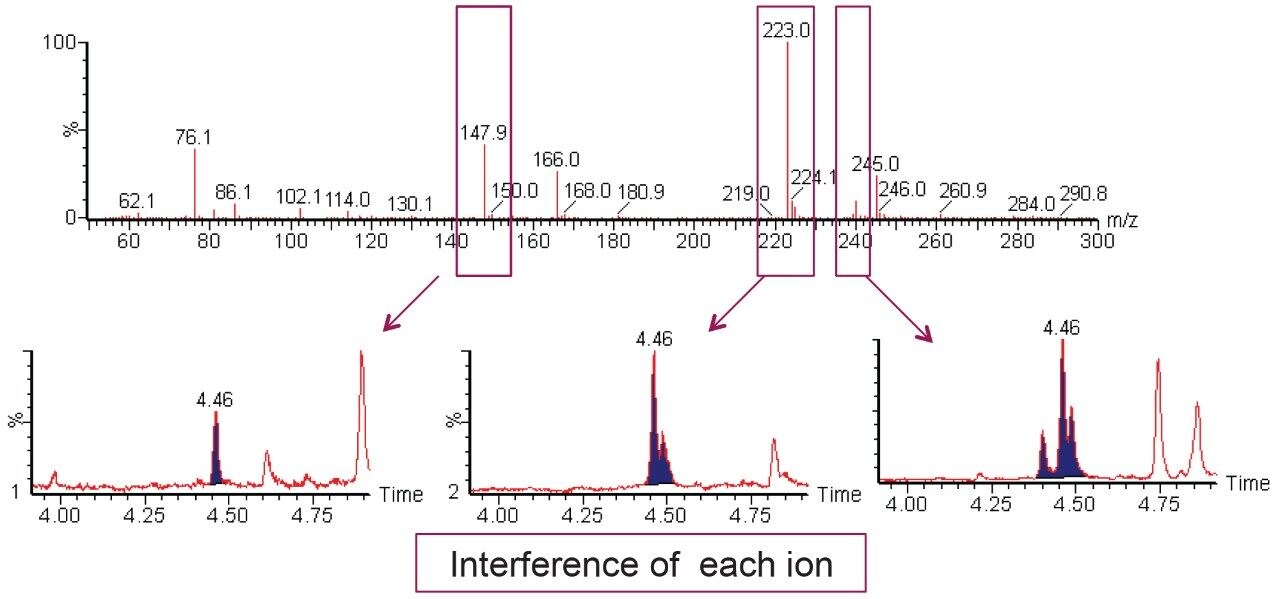
Optimal source conditions were investigated for carbamates before sample analysis. Ion masses monitored for each carbamate during the analysis were optimized and are listed in Table 1. Carbamate pesticides are generally labile and are prone to in-source fragmentation. For example, aldicarb sulfone forms three major ions (m/z 148, 223, and 240) during SIR (Selected Ion Recording) optimization. Ion mass m/z 148 was chosen for analysis because it is less affected by matrix interferences and provides good sensitivity to meet the performance requirements of the method, as shown in Figure 3.
Matrix effects were measured by comparing areas of post-spiked sample and solvent standard, where the spiked level was equal to 10 μg/kg, as shown in Table 2.
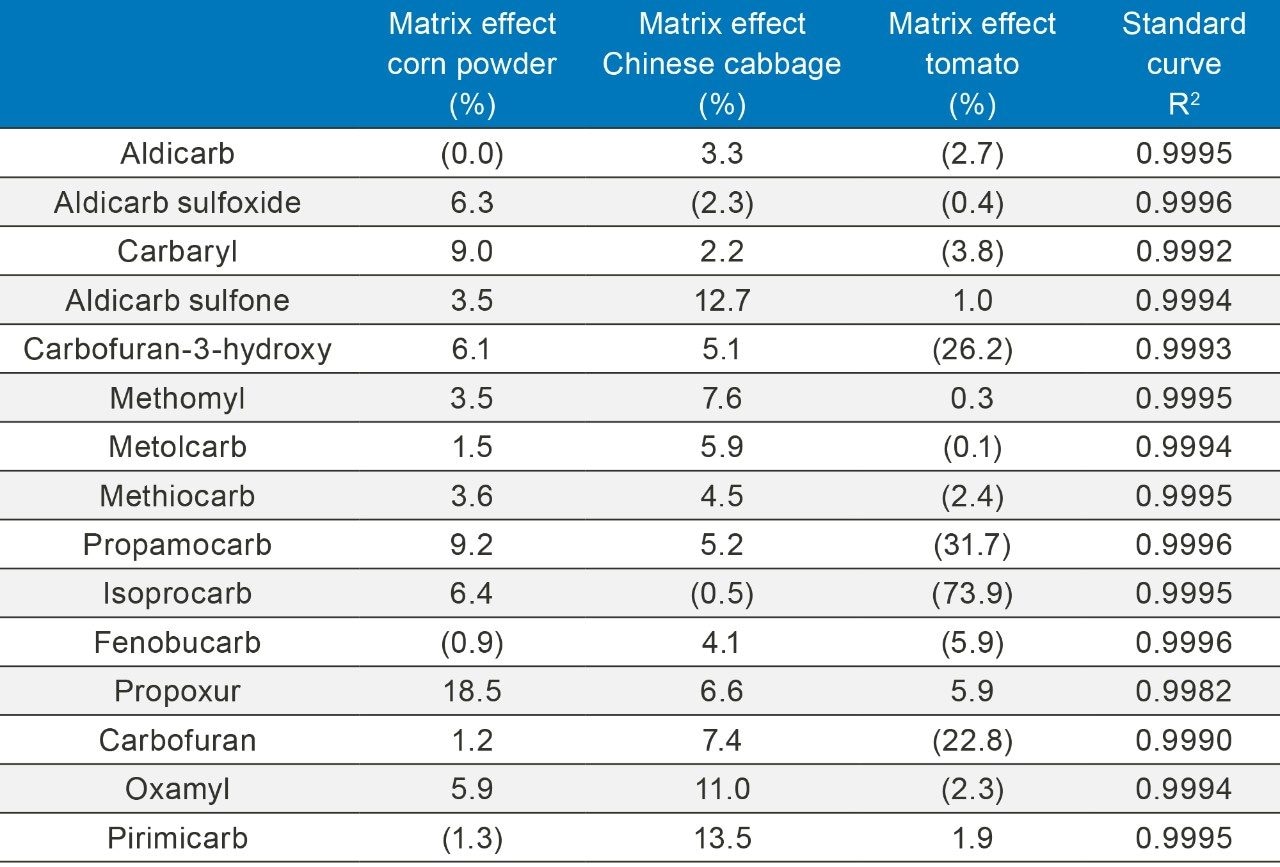
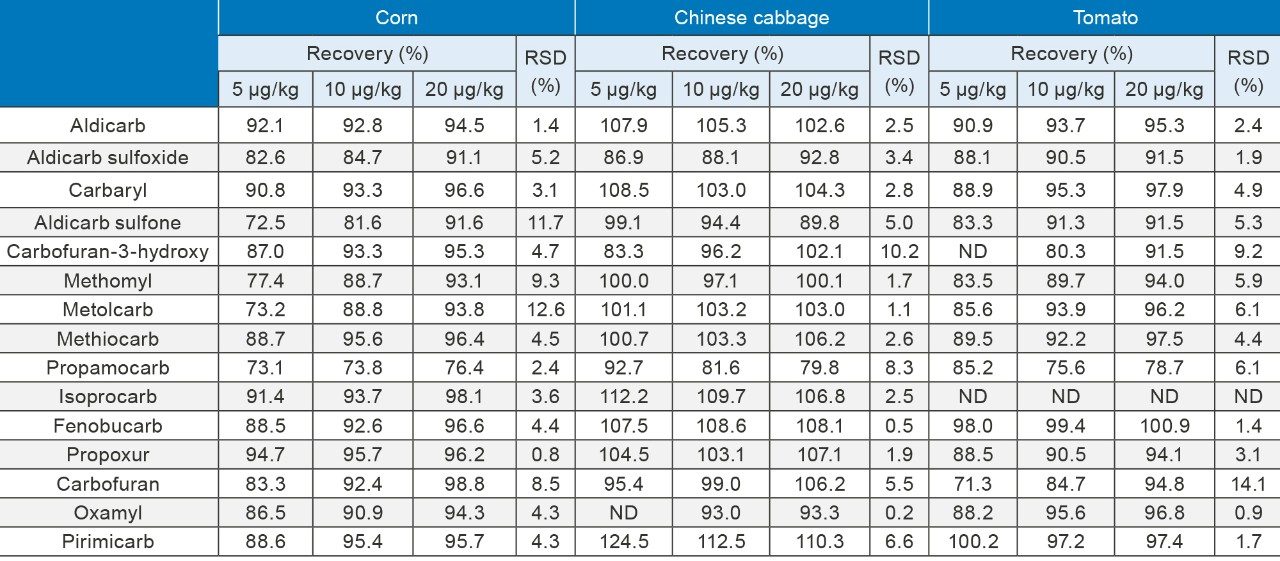
Analyte recovery was determined by spiking carbamate standards into the blank matrices (corn, cabbage, and tomato). The analytes were spiked at concentrations of 5.0 μg/kg, 10.0 μg/kg, and 20.0 μg/kg. Each level of spiking was repeated in three replicates. All of the samples were processed according to the method described previously. The concentrations were calculated using a matrix-matched calibration curve. The average recoveries and precision for each spiking level are listed in Table 3.
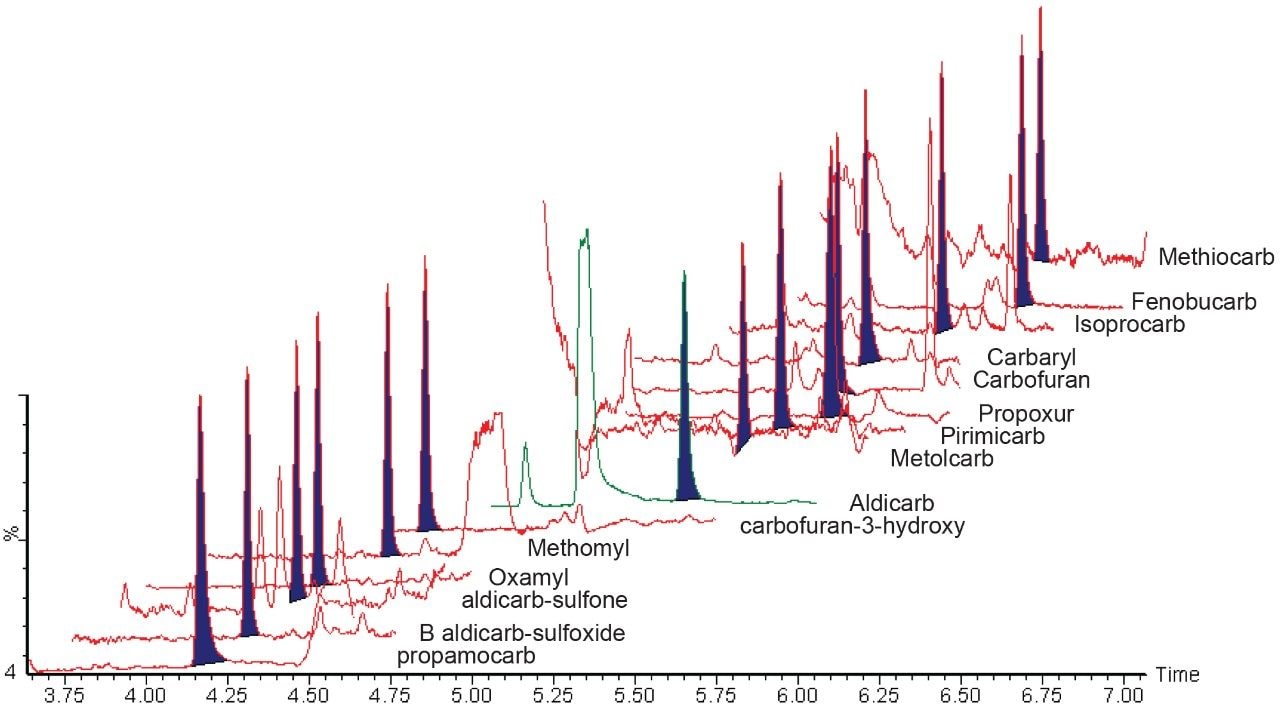
An example chromatogram was acquired and is shown in Figure 4, where the corn powder was fortified with carbamate standards at 5.0 μg/kg following the regulatory limits. Satisfactory sensitivity was observed for detection of each carbamate by UHPLC-MS.
720006215, February 2018