This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates to determine if the collisional cross section of a glycosylated peptide differs from a peptide of similar mass to allow separation and discrimination by an ion mobility spectrometry stage as part of an LC-MS analysis.
High Definition MSE (HDMSE) incorporates an IMS separation into both low and elevated collision energy scans and enables glycosylated peptide ions to be discriminated on the basis of their drift times.
Glycosylation is an important protein post-translational modification, involved in processes such as receptor recognition, protein solubilization, serum half life regulation, and conformation stabilization. Differences in the glycosylation patterns of particular proteins have been found to be indicators for certain cancers. Many biopharmaceutical proteins require glycosylation to be therapeutically safe and effective, and it is necessary to characterize and control the glycan structures displayed and control this glycosylation. As such, it would be useful to be able to discriminate, and therefore target, glycopeptides from native peptides during the course of an LC-MS experiment.
Ion mobility spectrometry separates ions by their collisional cross sections, which in turn are related to the size (mass) and shape of the ionized molecule, and the charge state of the ion. Attachment of a glycan to a peptide creates a branched structure rather than an extension of a linear peptide chain. Thus, it is conceivable that a glycopeptide could have a different collisional cross section than a native tryptic peptide, and therefore be distinguished by a different IMS drift time.
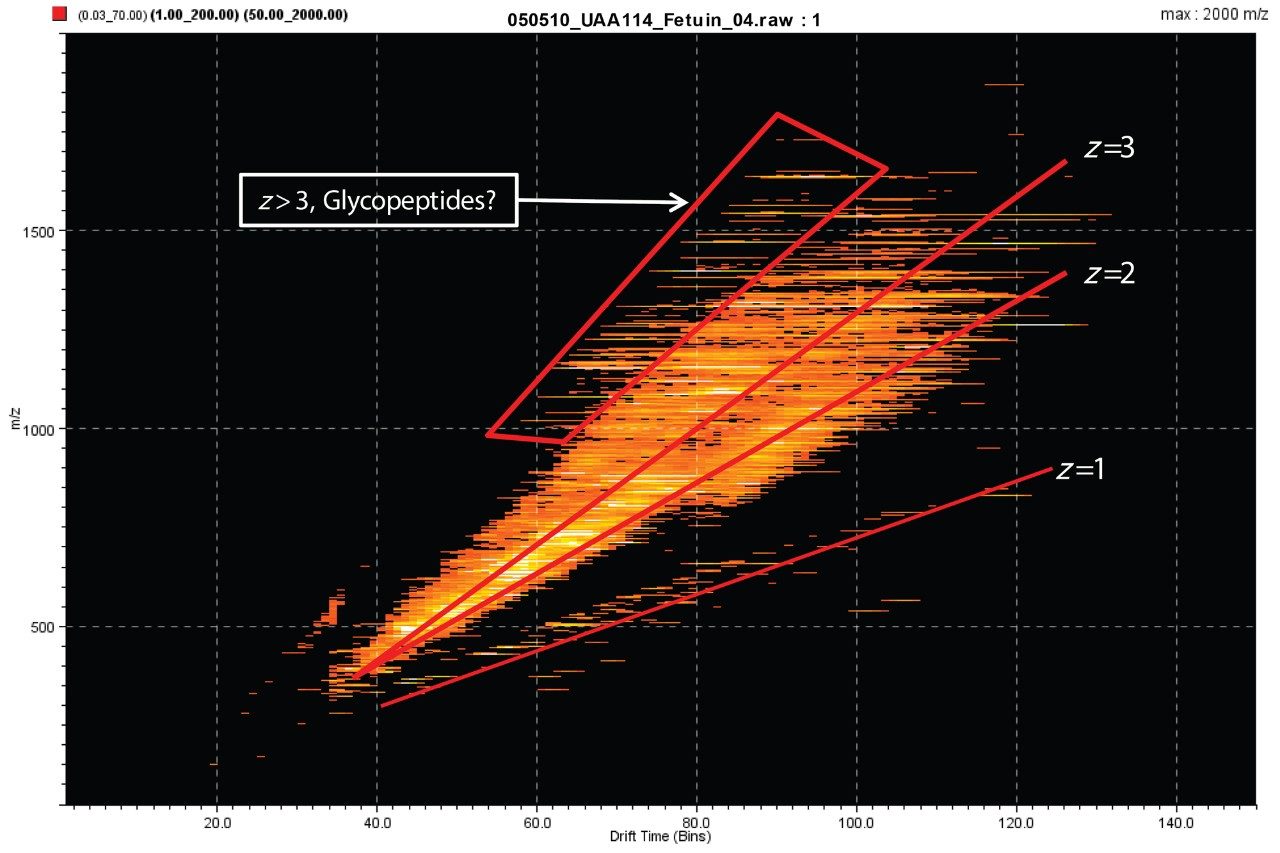
Bovine fetuin was reduced, alkylated, and trypsin-digested with the aid of RapiGest surfactant following recommended procedures. The resulting peptide and glycopeptide mixture was analyzed using a linear acetonitrile gradient on a nano LC (nanoACQUITY UPLC System) and HDMSE on a SYNAPT G2 Mass Spectrometer. The raw data are represented by the m/z and drift times of the ions in DriftScope, shown in Figure 1. The expected trend lines for charge states 1+, 2+, and 3+ are clearly visible, as is the boxed region of relatively high mass and charge state, which may represent the glycopeptides.
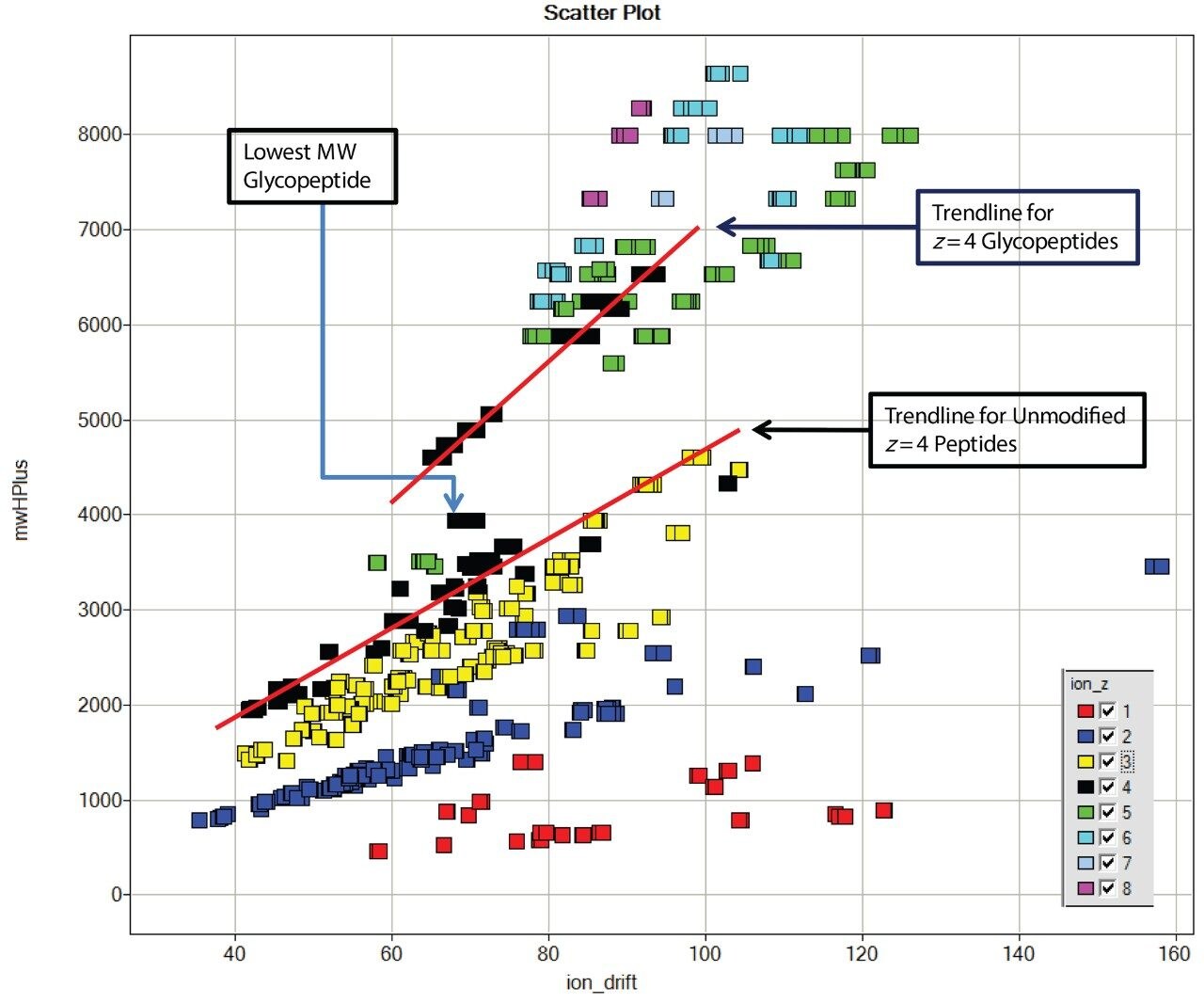
Data were processed with ProteinLynx Global SERVER v 2.4, which generated a table that includes the drift time, charge state, and calculated MH+ for each detected ion. This data is displayed using Spotfire DecisionSite (TIBCO), shown in Figure 2. The MH+/drift time trends for charge states 1+ (red), 2+ (blue), and 3+ (yellow) show the expected pattern. Two trend lines for charge state 4+ are visible. At lower MH+ values, the line follows that of the lower charge states, while for certain higher molecular weight peptides, corresponding to known glycopeptides, the trend line is different and the species have significantly shorter IMS drift times. The drift time for the lowest molecular weight glycopeptide (MH+ 3945) falls between. This is consistent with the hypothesis that the branched structure of a glycopeptide results in a shorter drift time than non-glycosylated peptides of same mass as the glycopeptides. Higher mass, higher charge state ions of larger glycopeptides cluster around this trend line as a “cloud” separate from the bulk of the peptide ions.
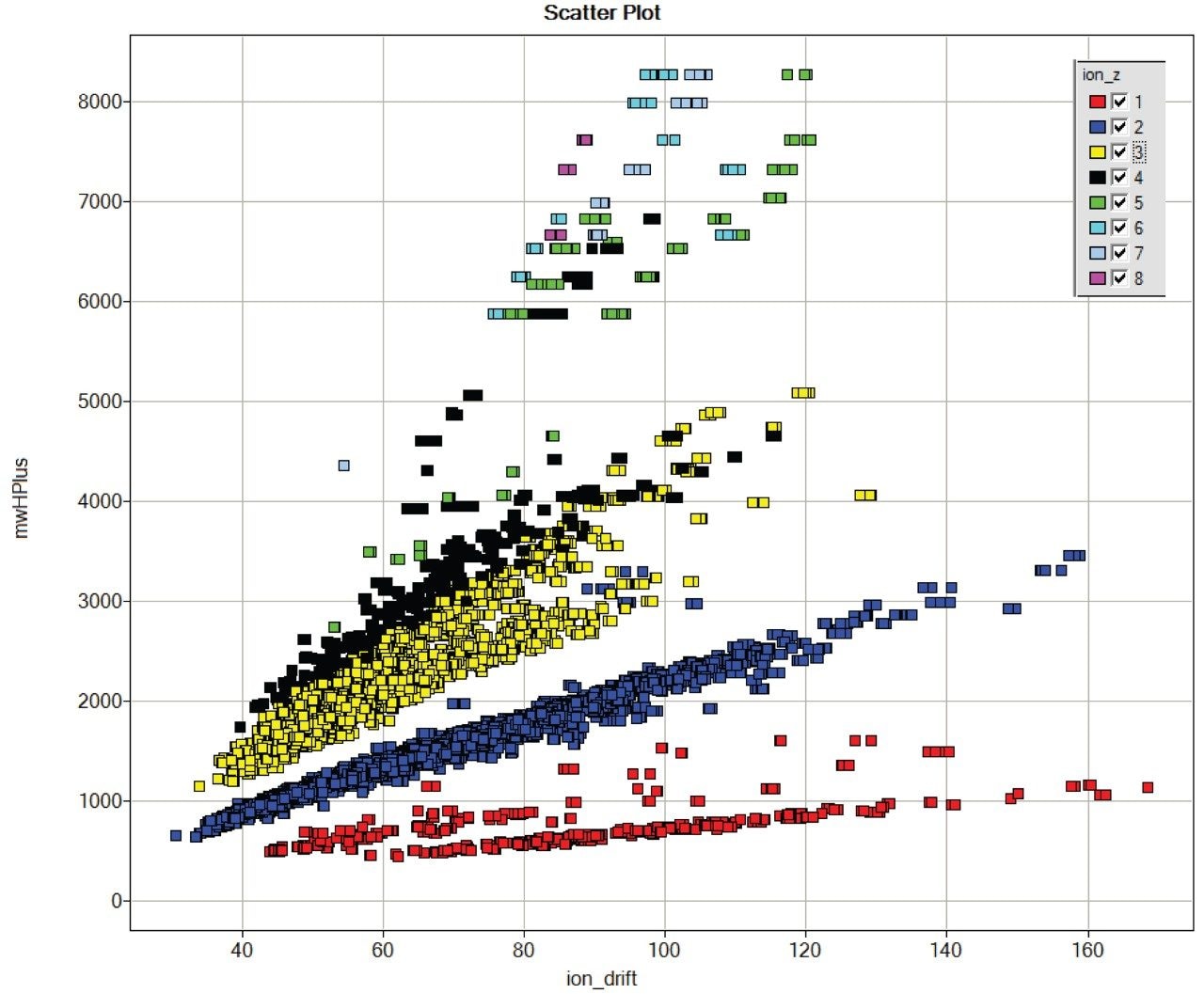
The same segregation of glycopeptide ions is seen when fetuin is added to a digest of E. coli proteins, shown in Figure 3.
Addition of a glycan to a peptide often alters its collisional cross section in a fashion that facilitates segregation from unmodified peptides by ion mobility spectrometry. This in turn facilitates characterization of glycosylated peptides in complex protein digests. Only HDMSE provides this information as part of a proteomics workflow. The addition of ion mobility separation to LC-MSE data collection with the SYNAPT G2, along with its enhanced mass resolution, makes this instrument a unique tool for the study of protein glycoslyation.
720003882, March 2011