
In this application note we describe a new method for sample preparation that involves the use of a new MALDI target plate.
Matrix assisted laser desorption/ionisation-time of flight mass spectrometry (MALDI-Tof MS) is a widely used and highly specific technique for the analysis of proteins and peptides. And it’s now a well-established tool in the biochemists' toolbox. Technologies that facilitate the automated spot excision of gel electrophoresis-separated proteins and robotic in-gel tryptic digestion prior to MALDI analysis of the resulting peptide mixtures are now commonplace. These high throughput, integrated protein analysis systems, such as the ProteomeWorks System, have proven extremely successful for the analysis of large numbers of proteins from a wide range of biological organisms.
MALDI is a robust technique that generally has a high tolerance of salts and contaminants, however removal of these impurities prior to analysis significantly improves the data quality, the number of peptides observed and can vastly improve the confidence with which proteins are identified.
In this application note we describe a new method for sample preparation that involves the use of a new MALDI target plate. The Waters MassPREP PROtarget Plate is coated with a thin film of hydrophobic polymer and comprises 96 sample wells (and 24 for near-point lockmass) with a hydrophobic center, designed to effectively bind peptides to the plate. This allows repeated sample washing to remove contaminants such as salt and detergents, prior to addition of the matrix solution and analysis by MALDI-Tof MS.
This new sample preparation procedure has been compared against use of a stainless steel plate for the analysis of low abundance proteins. The MassPREP PROtarget Plate is shown to have several advantages over the stainless steel plate:
Standard proteins: Bovine serum albumin, Phosphorylase B and Myoglobin (Sigma, St Louis, MO) were dissolved in water (1 mg/mL) and all three proteins combined to give an equal concentration solution of 333 fmol/μL per component. Sample was loaded (1 μl, 5 μl, 10 μl, and 15 μl) and separated on a 1-D Criterion Precast gel (Bio-rad, Hercules, CA). After separation, the gel was stained using BioSafe Blue Coomassie stain and the 1D bands were excised manually and split into 2 or 4 gel pieces. The gel pieces were put in a NUNC 96-well microtiter plate.
Gel pieces with total sample loading of 83, 416, 833, and 1250 fmol for BSA and Phosphorylase B, and 166, 416, 833, and 1250 fmol for Myoglobin were produced.
The samples were processed using the Waters MassPREP Station liquid handling robot. The control software of the MassPREP station (Digest SPP 2.0 bis) allows destaining, reduction, alkylation, digestion, extraction and spotting of the samples onto a stainless steel target and the MassPREP PROtarget Plate. Automated preparation of samples on the MassPREP PROtarget Plate is simple and consists of five steps:
All data were acquired in positive ion mode on a Waters Micromass MALDI-Tof Mass Spectrometer. The mass spectra were collected in automated fashion, over a m/z range of 800–3500 in reflectron mode. Calibration was performed by a separate acquisition of a peptide mixture.
Waters ProteinLynx Global SERVER 2.0 Bio- Informatics Software was used for data processing and database searching.
During the data processing step, a near-point lockmass was applied, (Glu1)-Fibrinopeptide B. The mass spectra were smoothed, background subtracted and monoisotopic peaks were identified through the use of the MaxEntLite algorithm.
Data were converted to XML format and searched against the Swiss-Prot database v40.
Results were obtained from all the gel digest samples using both the new MassPREP PROtarget Plate and the stainless steel target plate. Two parameters were chosen to compare and contrast the obtained results. The first of these is the percent sequence coverage of the protein identified through database searching. For each sample either two or four replicates have been averaged. Secondly, the average signal intensity obtained from the peptide mass fingerprint has been calculated for comparative purposes. The average intensity was defined in this case as the absolute signal response obtained from the three major peaks (for each protein).
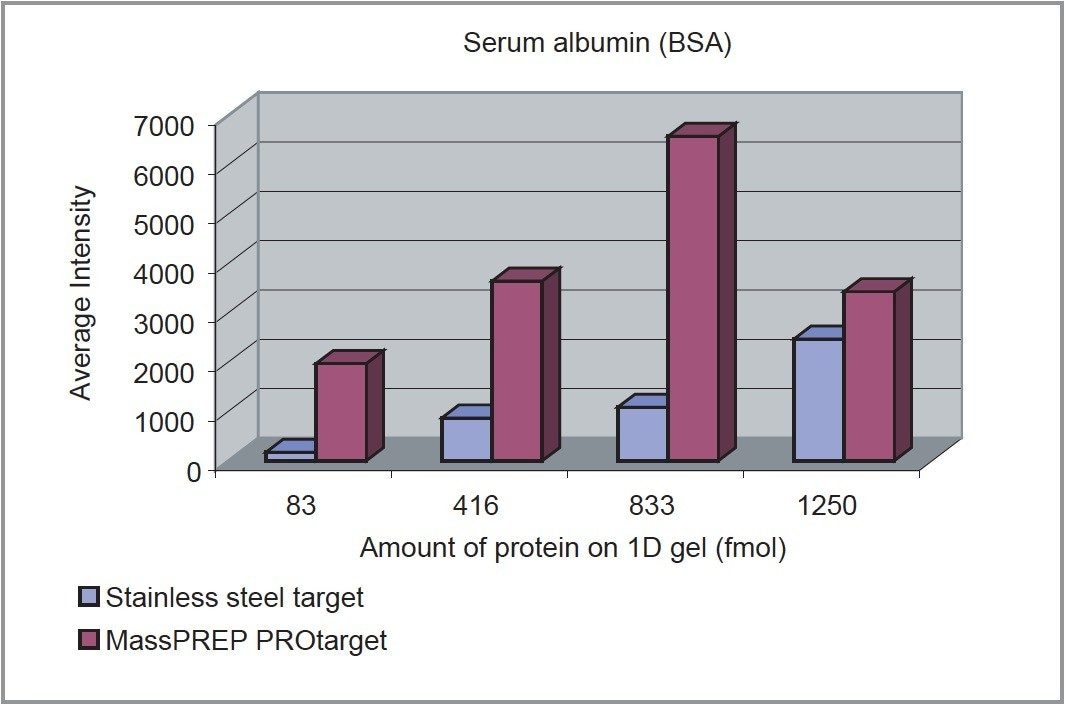
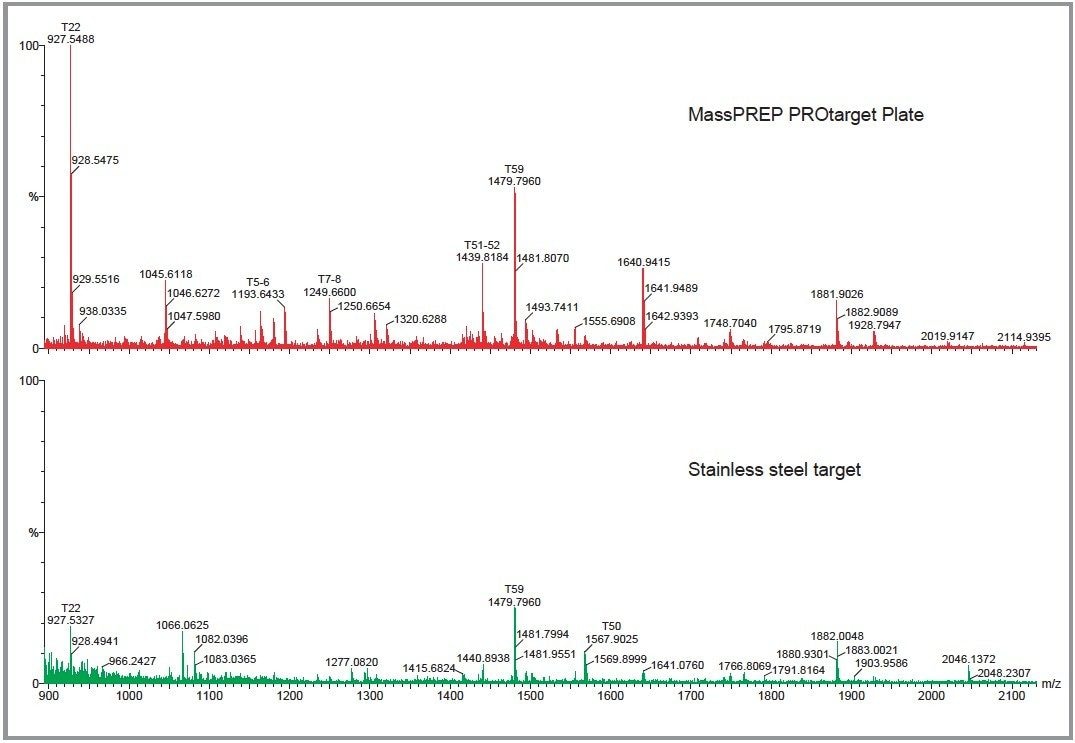
The results obtained are summarized in Table 1. Both the intensity of the peaks and the percent sequence coverage are dramatically improved using the MassPREP PROtarget Plate in comparison to the stainless steel target. Additionally, the average intensity of the peaks is greater in all cases using the new target. Analysis of the 1250 fmol BSA sample resulted in an average intensity decrease in comparison to the analysis of BSA at the 833 fmol level (Figure 1), probably due to the overloading of the sample on the ActiveWell. This is not observed in the case of the other two proteins, where the average intensity increased with the sample loading.
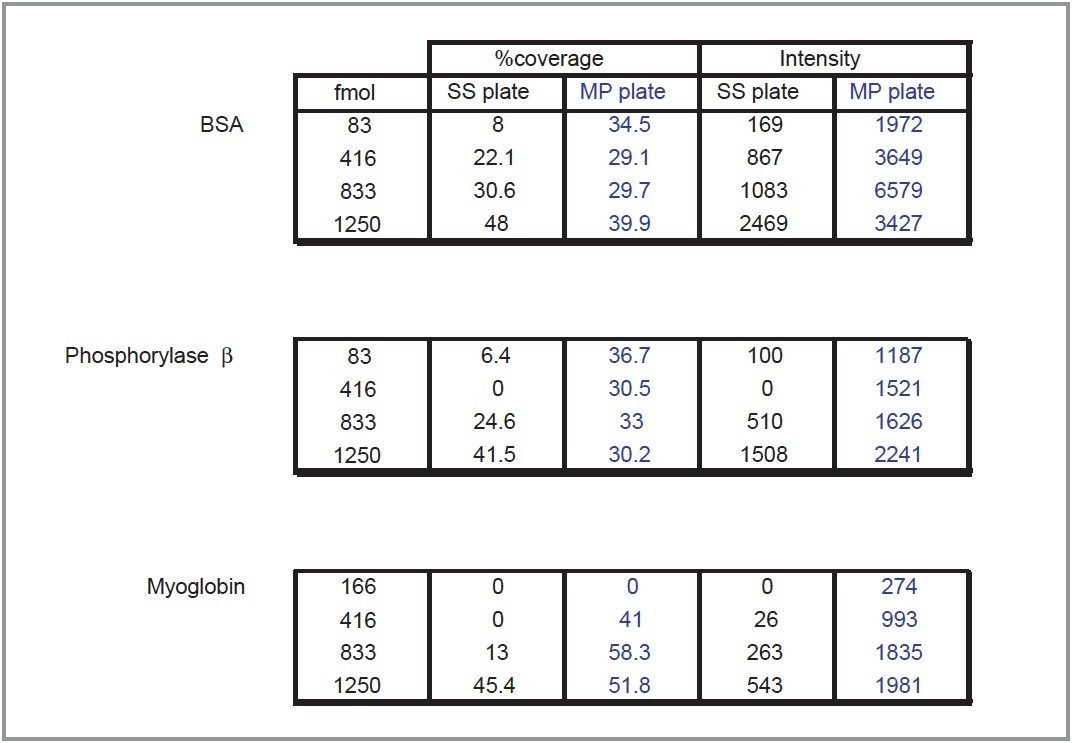
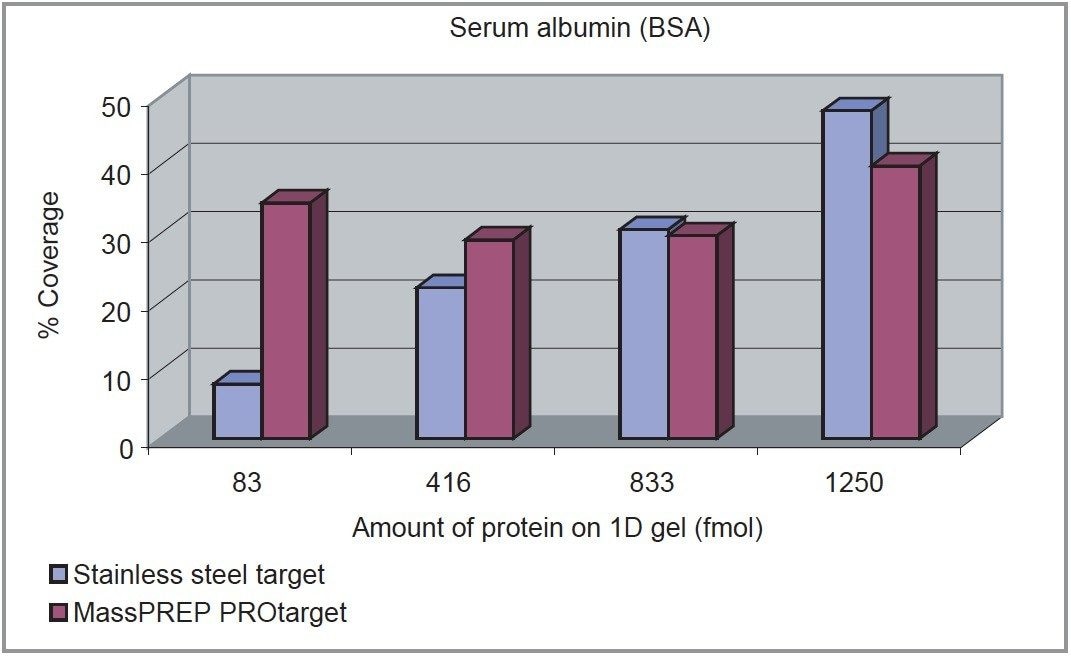
In the case of the new MassPREP PROtarget Plate, the percent sequence coverage is consistently high even at the lowest concentration tested (83 fmoles). In comparison, the stainless steel target’s sequence coverage is considerably lower at 83 fmoles and increases linearly to the 1250 fmole level.
As illustrated in Figure 2, at high sample concentrations, the percent sequence coverage is slightly better using the stainless steel target. This can be explained by the suppression of low intensity peptides by high intensity species. However, use of the MassPREP PROtarget Plate has shown that all three proteins can be identified, with high percent sequence coverage at low sample loadings on 1D gels. There are clearly significant advantages of using this new technology, particularly with samples obtained from 1D or 2D gel separations where the protein concentration is low.
720000740, October 2003