For research use only. Not for use in diagnostic procedures.
A robust, reliable bioanalytical method for the absolute quantification of twenty amino acids and the relative quantification of a further eighteen amino acids in mammalian urine has been developed and evaluated. The method had an analysis time of 7.5 minutes per sample. This allows the analysis of two, 96-well, microtitre plates of samples in a 24 hour time period. The method was found to be valid over the physiologically important range of 0.2–200.0 μMol.
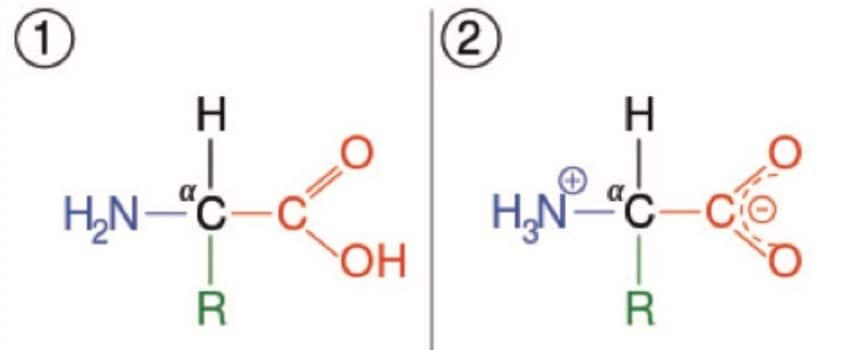
Amino acids play a critical role in mammalian biochemistry, forming the simple building blocks of proteins, acting as neurotransmitters in biosynthesis and are essential in lipid transports, to name but a few. The rapid and accurate quantification of amino acids is critical to understanding the underlying biochemistry of multiple physiological and disease states. Previous methodologies have employed either ion exchange chromatography followed by derivatization with fluorescence detection or sample derivatization followed by analysis by LC-UV or LC-Fluorescence. However, both of these approaches are time consuming and require complete chromatographic resolution of the amino acids from other amine-containing compounds, so are not always suitable for the analysis of biological fluids. Here we present a bioanalytical method for the rapid, simple, quantification of amino acids by UPLC-MS/MS for research use.
|
LC System: |
ACQUITY UPLC I-Class |
|
Detector: |
Xevo TQ-S micro |
|
Vials: |
Maximum Recovery vials |
|
Column: |
ACQUITY UPLC HSS T3 1.8 μm, 150 x 2.1 mm |
|
Column temp.: |
45 °C |
|
Sample temp.: |
Room temperature |
|
Injection vol.: |
2-μL |
|
Flow rate: |
0.6 mL/min |
|
Mobile phase A: |
Water + 0.1% formic acid |
|
Mobile phase B: |
Acetonitrile + 0.1% formic acid |
|
Gradient: |
Maintained at 4% B for 0.5 min; increasing to 10% B at 2.5 min; increasing to 28% B at 5 min, increasing to 95% B at 5 .1 min; reverting to 4% B at 6.2 min for a 1.3 min re-equilibration see Table 1 |
|
MS system: |
Xevo TQ-S micro |
|
Ionization mode: |
Positive |
|
Acquisition range: |
MRM mode |
|
Capillary voltage: |
1.5 kV |
|
Collision energy: |
Compound specific, see Table 2 |
|
Cone voltage: |
Compound specific, see Table 2 |
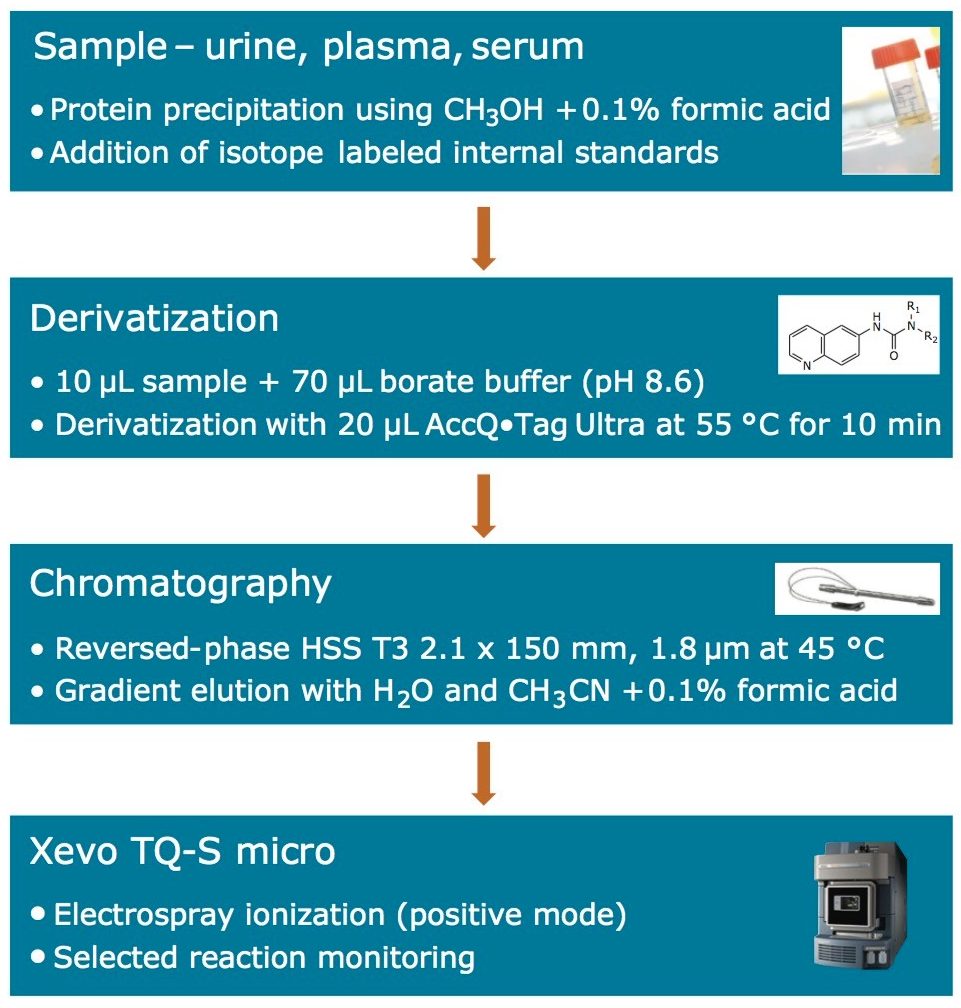
The sample preparation procedure employed for the analysis of the amino acids is shown below in Figure 2. A 50-μL aliquot of the samples and standards was vortex mixed with 150-μL of methanol, to precipitate protein. A 10-μL aliquot of the resultant sample was then transferred to a sample tube for derivatization according to the process defined in Figure 2.
|
Time (min) |
Flow rate (mL/min) |
%B |
|---|---|---|
|
0.0 |
0.6 |
4 |
|
0.5 |
0.6 |
4 |
|
2.5 |
0.6 |
10 |
|
5.0 |
0.6 |
28 |
|
5.1 |
0.6 |
95 |
|
6.1 |
0.6 |
95 |
|
6.2 |
0.6 |
4 |
|
7.5 |
0.6 |
4 |
Table 1. Chromatographic gradient table.
The bioanalytical method was subjected to a one day validation according to the FDA guidelines for bioanalytical method validation. The samples employed are shown below:
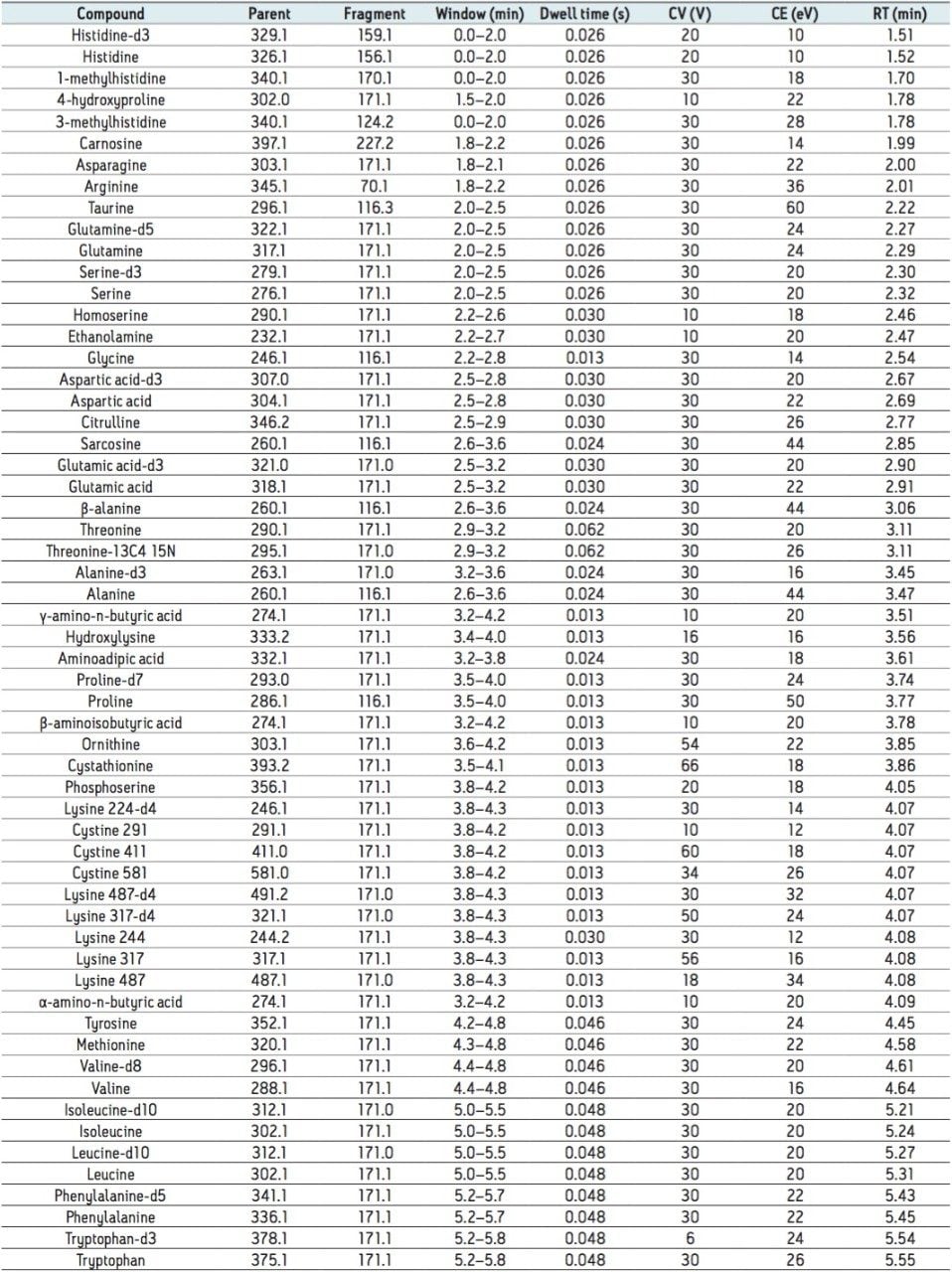
The amino acids analyzed in this study are listed in Table 3. The twenty proteinogenic amino acids were subjected to validation for absolute quantification using stable isotope labeled internal standards. Those additional eighteen amino acids, for which no stable isotope labeled internal standard was used, were subjected to relative quantification. The stable isotopes employed for each amino acid is listed in Table 4.
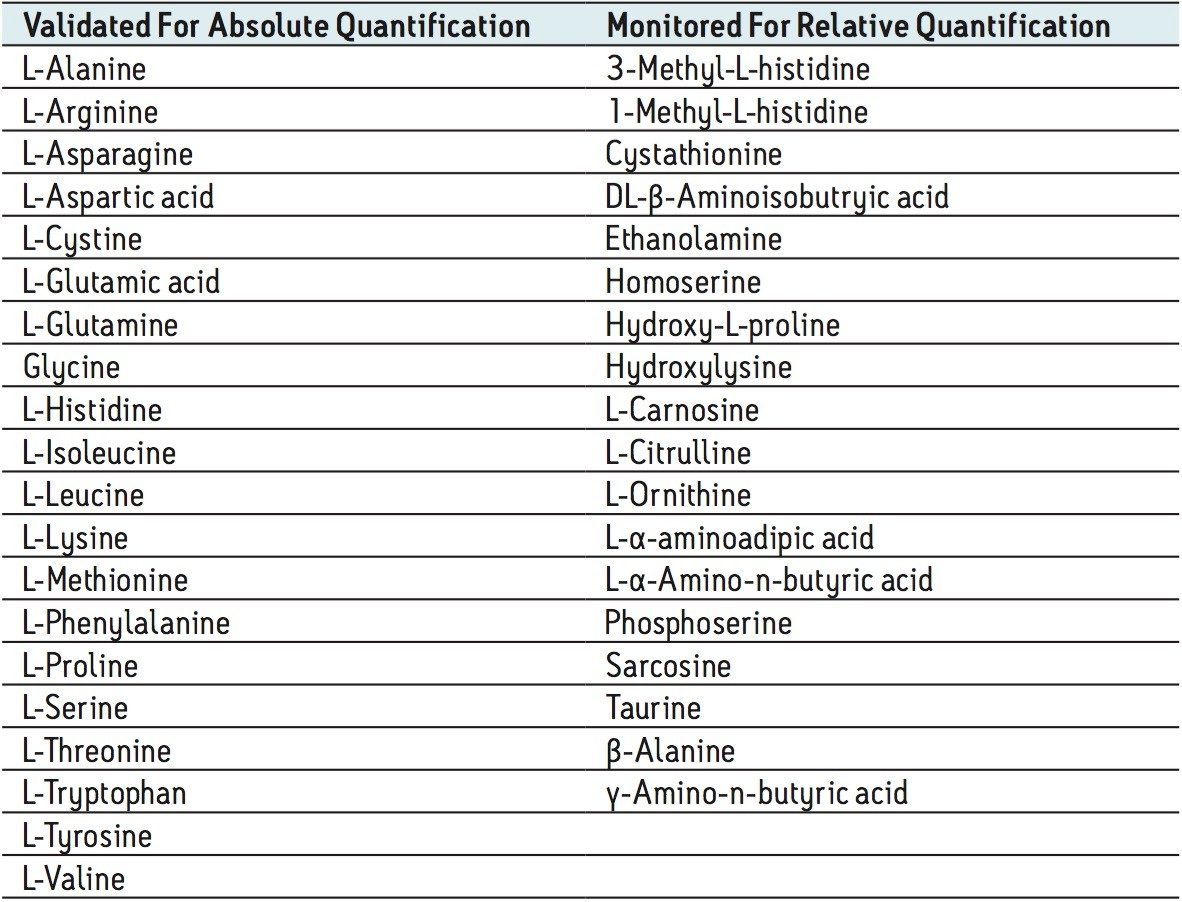
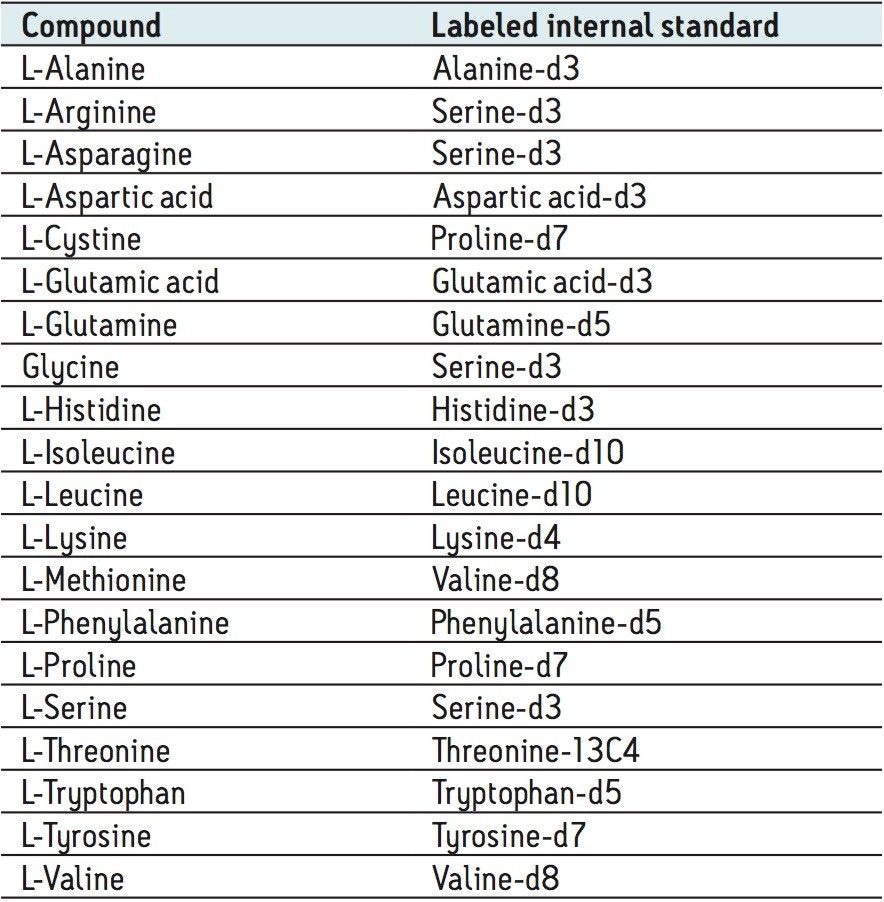
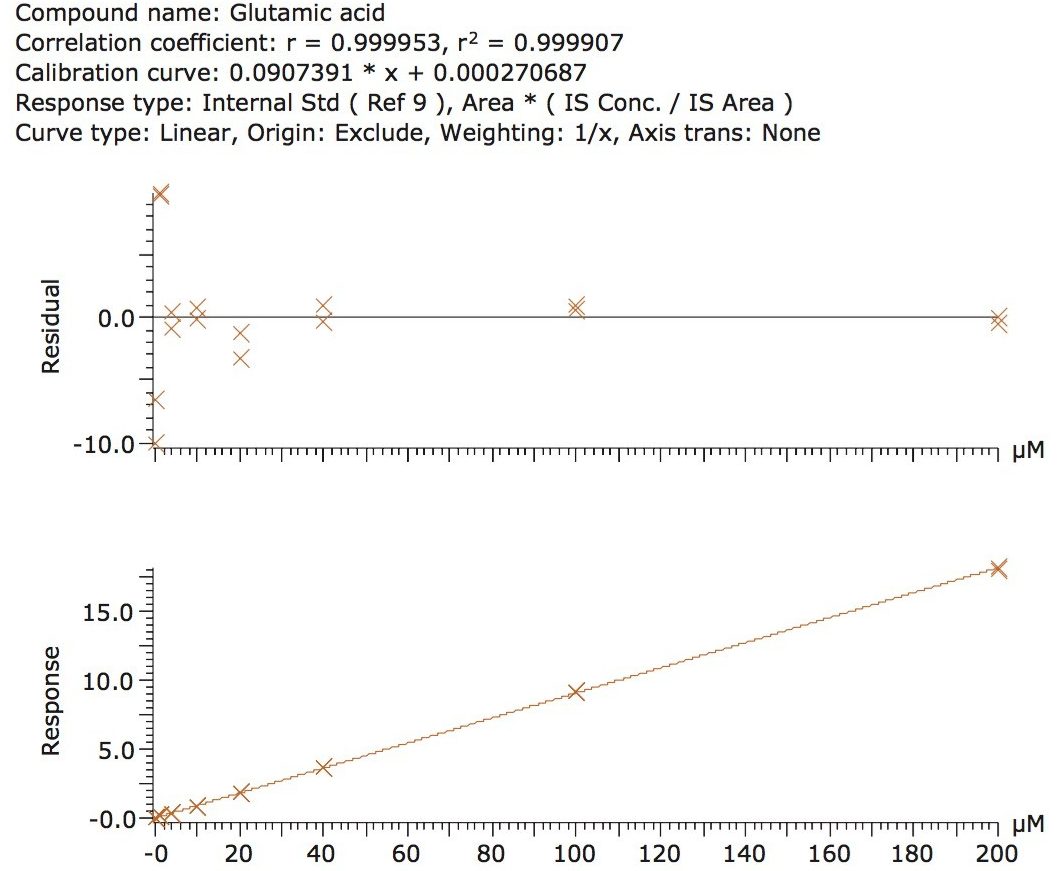
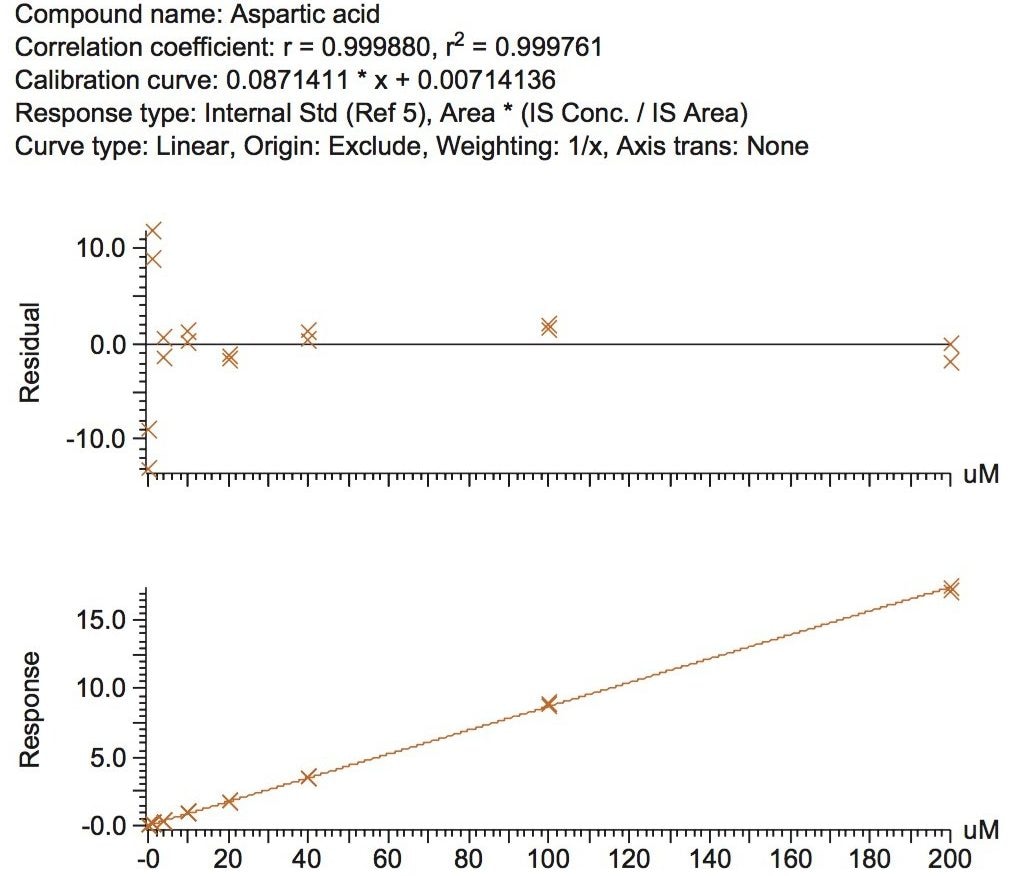
The calibration line was prepared from a Sigma Aldrich physiological, amino acid standard (acidics, basics, neutrals) by spiking into 50/50 CH3OH/H2O. The calibration curve was generated over three orders of magnitude, covering the physiological range of 0.2–200.0 µM. The QC samples were prepared from a separate Sigma Aldrich physiological amino acid standard.
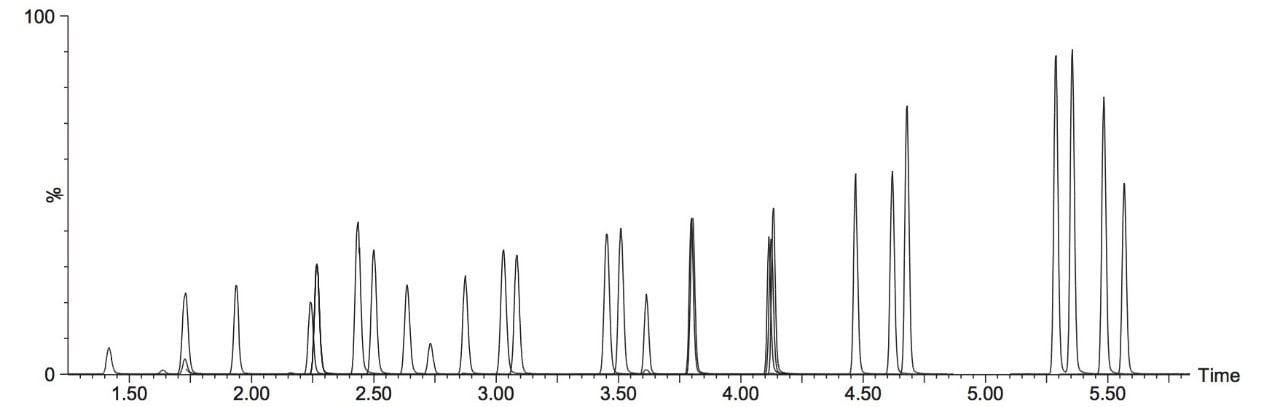
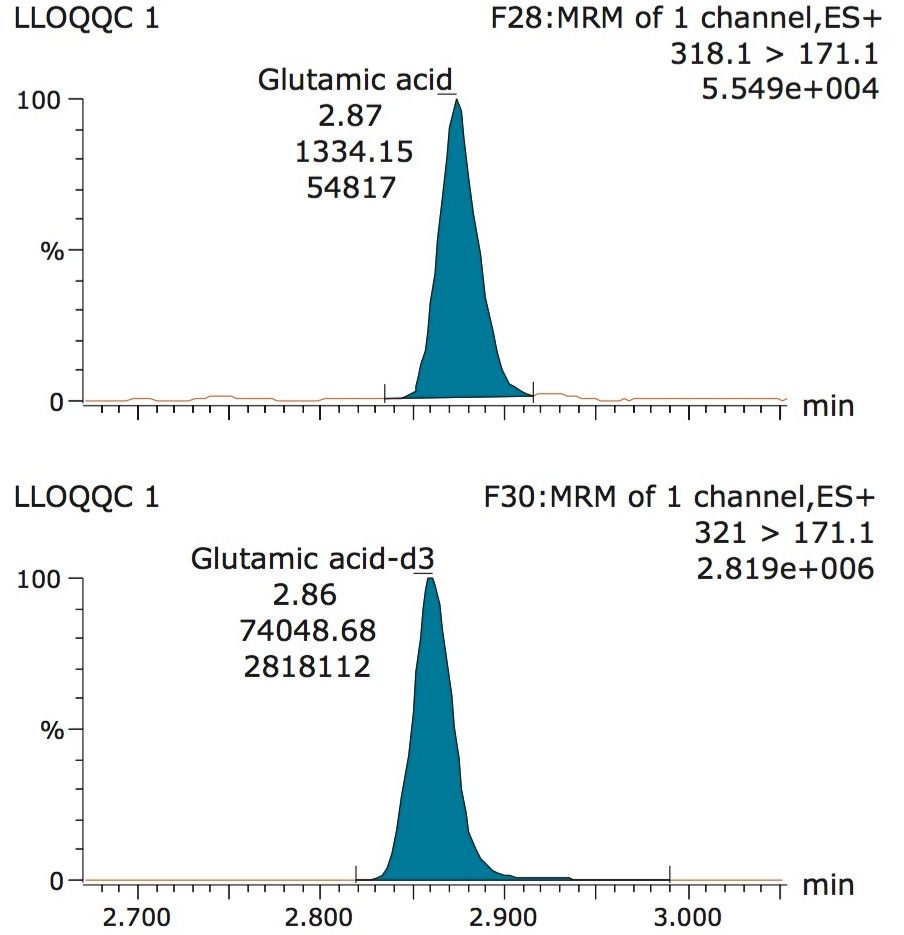
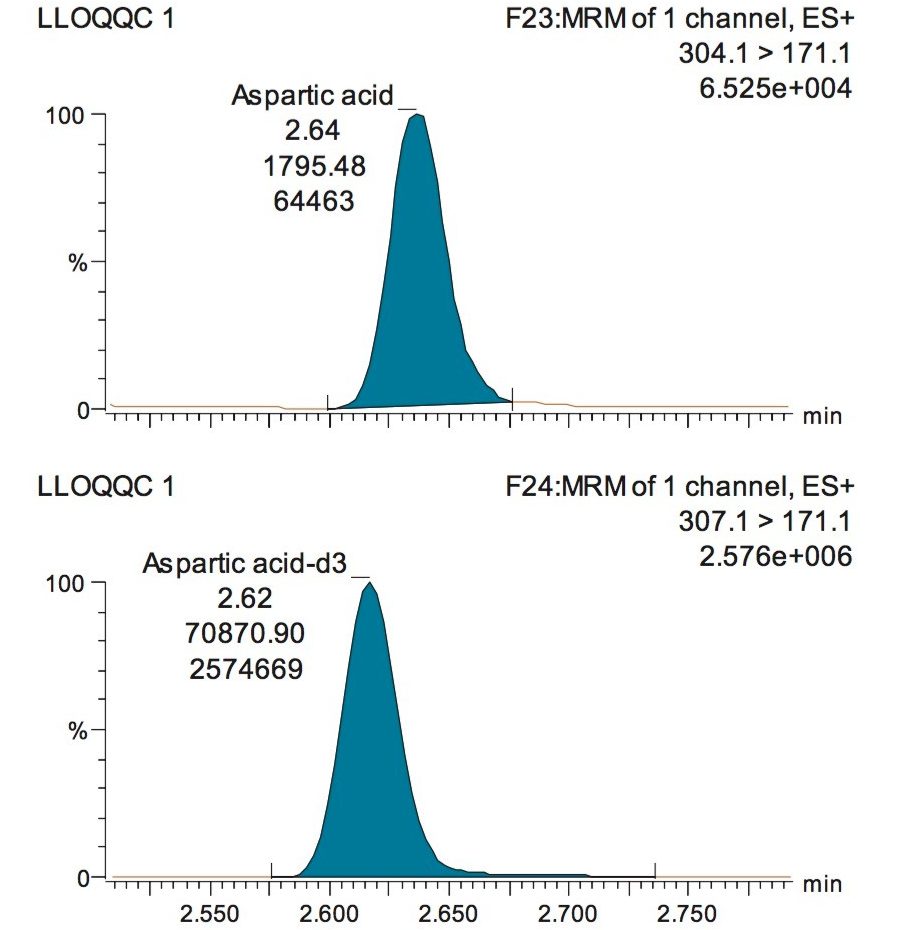
A typical separation obtained from the amino acid standard mix is shown in Figure 3. The data displayed illustrates the separation obtained for the amino acids and the throughput of the method. The peak shape obtained from the chromatography was excellent with a peak width at the base of approximately 3 seconds. The method was shown to be reproducible and reliable with no retention time drift. The Xevo TQ-S micro is equipped with a new novel, state of the art, ion transfer optics which allows more ions to be sampled from the source and transferred to the analyzer. The StepWave ion guide in the Xevo TQ-S micro is designed to cope with the challenges of the modern laboratory that are produced by high sample throughput and difficult matrices. Neutrals and gas load are passively removed for enhanced transmission with the ions actively transferred into the mass analyzer, improving sensitivity and robustness.
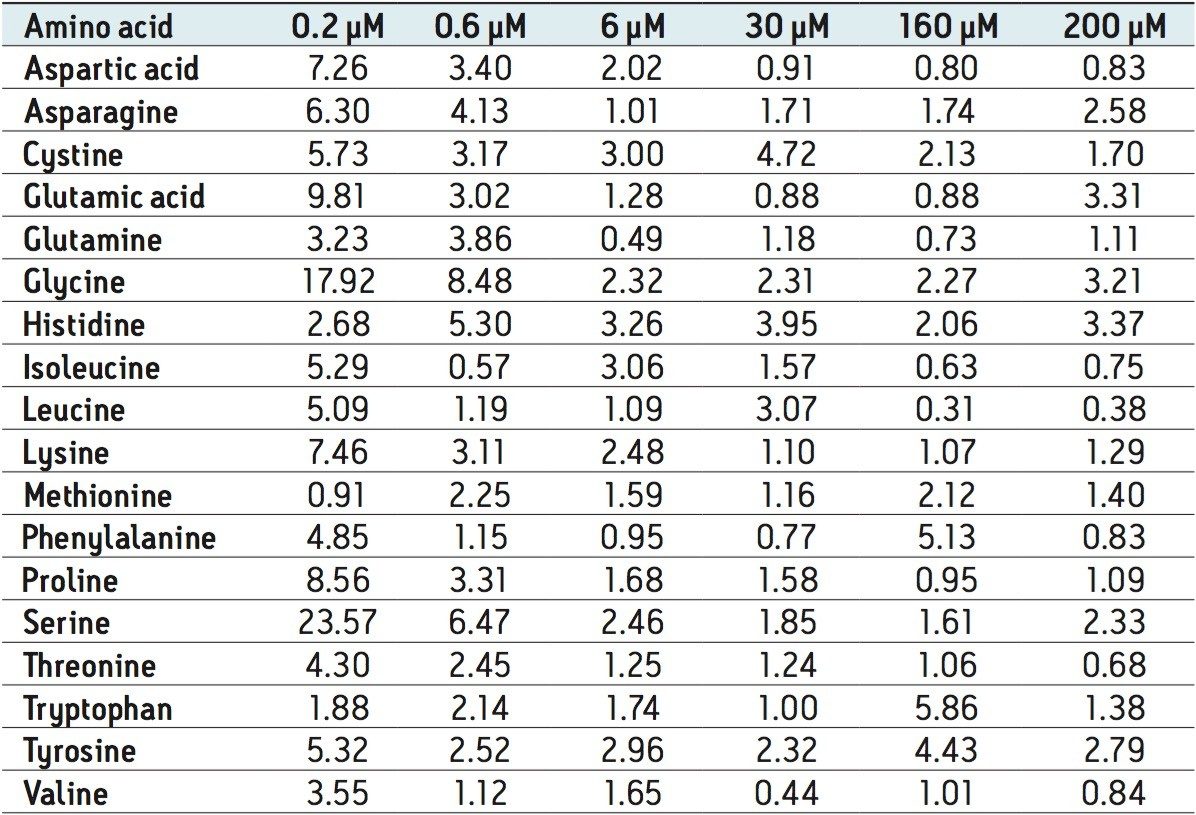
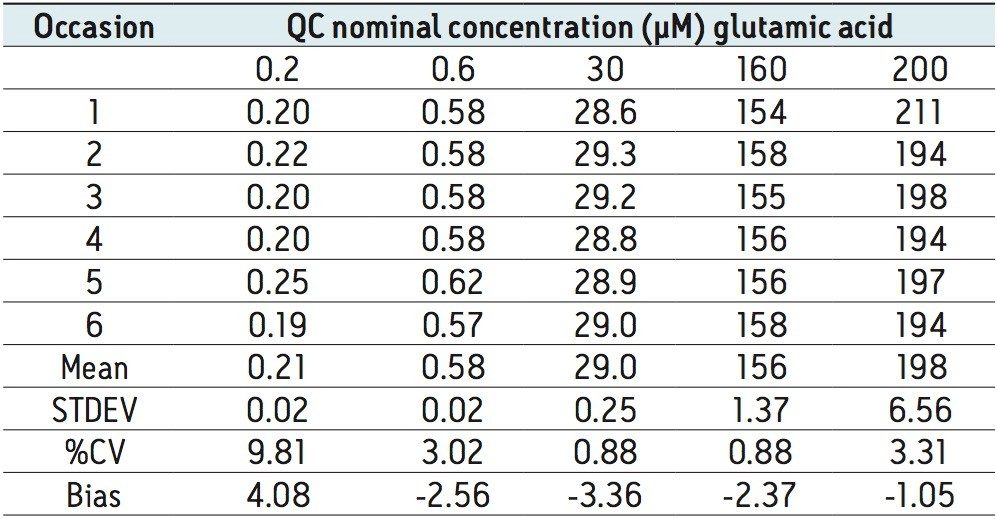
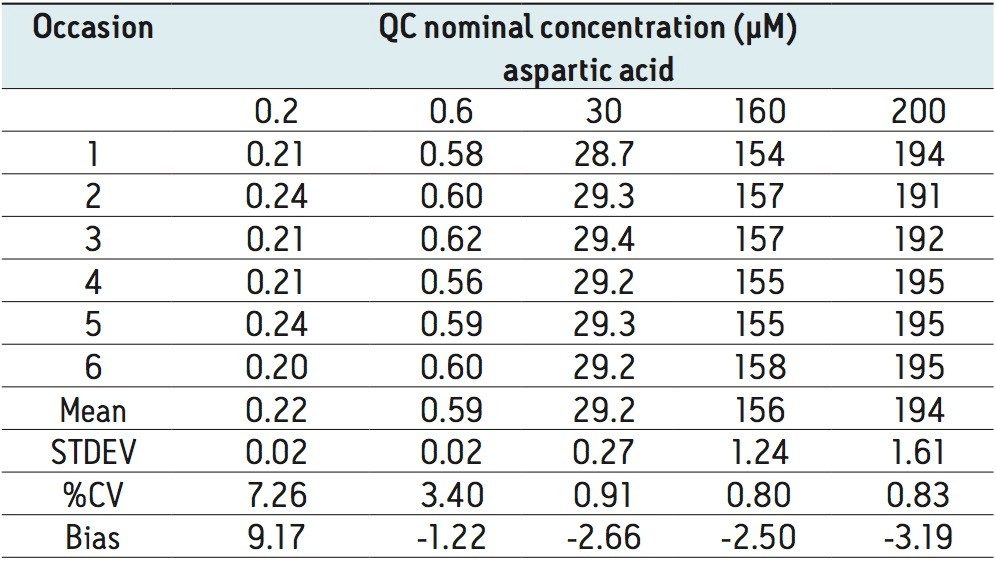
The method validation was carried out according to FDA guidelines for bioanalytical methods. The method is not validated for clinical diagnostic use. The resulting amino acid data was processed and quantified using Waters® MassLynx TargetLynx application manager employing internal standard calibration and 1/x weighting. A summary of the QC data for each amino acid is listed below in Table 5. The data obtained for the quantification of each amino acid was acceptable for routine quantification.
The validation results as well as example chromatograms and calibration lines for the amino acids glutamic acid and aspartic acid are shown in Figures 4–7 and Tables 6 and 7. Here we can see that the methodology demonstrated acceptable peak shape and signal to noise ratio at the lowest level of quantification. The method demonstrated excellent bias and precision for every amino acid.
A robust, reliable bioanalytical method for the absolute quantification of twenty amino acids and the relative quantification of a further eighteen amino acids in mammalian urine has been developed and evaluated. The method had an analysis time of 7.5 minutes per sample. This allows the analysis of two, 96-well, microtitre plates of samples in a 24 hour time period. The method was found to be valid over the physiologically important range of 0.2–200.0 µMol. The chromatography was reproducible and reliable with no retention time drift detected for any of the amino acids. This data demonstrates that LC-MS/MS provides an attractive, viable, and alternative bioanalytical method to traditional modes of amino acid analysis, providing fast and accurate quantification.
720005189, September 2016