In this application note we describe the use of a Q-Tof Premier in combination with a nanoACQUITY UPLC System for the detection of low-level tryptic digests.
The use of LC-MS/MS to qualitatively characterize enzymatic protein digests is now standard and widespread throughout protein laboratories. This method has been shown to work well with mixtures of low and medium complexity, while highly complex protein samples are often separated using an extra stage of chromatography prior to analysis via LC-MS/MS.
The mass spectrometer is normally operated in the Data Directed Analysis (DDA) mode, where the instrument automatically selects peptides for MS/MS, based upon their intensity and charge state, as they elute from the HPLC column.
In this application note we describe the use of a Q-Tof Premier in combination with a nanoACQUITY UPLC System for the detection of low-level tryptic digests.

Samples of the MassPREP Digest Standards, Enolase, and Bovine Serum Albumin (Waters Corp, Milford MA) were analyzed on the Q-Tof Premier nanoACQUITY UPLC System. Stock solutions at a concentration of 1 pmol/μL were prepared by adding 1 mL of aqueous 0.1% formic acid to the lyophilised powder. Samples were then diluted to the desired concentration for injection using 0.1% formic acid. The nanoACQUITY sampler manager was programmed to inject 2 μL in full loop mode.
Samples were injected, and separated, on a nanoACQUITY System configured in trapping mode, (with a trapping and analytical column). The trapping column (180 μm ID x 20 mm), was packed with Symmetry C18 5 μm, while the analytical column (75 μm ID x 100 mm) was packed with Atlantis dC18 3 μm. Solvent A was aqueous 0.1% formic acid with solvent B acetonitrile + 0.1% formic acid.
Trapping was performed for 3 minutes using the Binary Solvent Manager operating at a flow rate of 5 μL/min and at a composition of 3% B. The gradient then changed the composition of the solvent B from 3% B to 40% B over 30 minutes with a wash at 95% B for 2 minutes. The columns were then re-equilibrated for 30 minutes prior to the next injection. The nanoACQUITY provides direct flow to the nano-scale column, without flow splitting. In these experiments a flow rate of 250 nL/min was used to elute the peptides from the analytical column.
All analysis was carried out using the Waters Micromass Q-Tof Premier mass spectrometer operating in the V-Optics mode. The Q-Tof Premier was operated in Data Directed Analysis mode with an MS survey integration time of 1 second.
The mass spectrometer was programmed to switch into the MS/MS mode on a single precursor ion, when a doubly or triply charged ion exceeded a threshold of 6 counts/sec. MS/MS was performed with an integration time of 2 seconds per precursor, up to a maximum of 12 seconds for each selected ion.
The column eluent was coupled directly to a NanoLC sprayer, fitted to the NanoLockSpray source of the mass spectrometer. In the reference channel of the NanoLockSpray source a 300 fmol/μL solution of Glu Fibrinopeptide B was continually delivered by the Auxiliary Solvent Manager of the nanoACQUITY System to provide a lock mass ion at m/z 785.8426. The ASM was operated at a flow rate of 0.5 μL/min.
The mass spectrometer was calibrated, prior to the analysis, using a mixture of CsI and NaI, over the m/z range 50–1990.
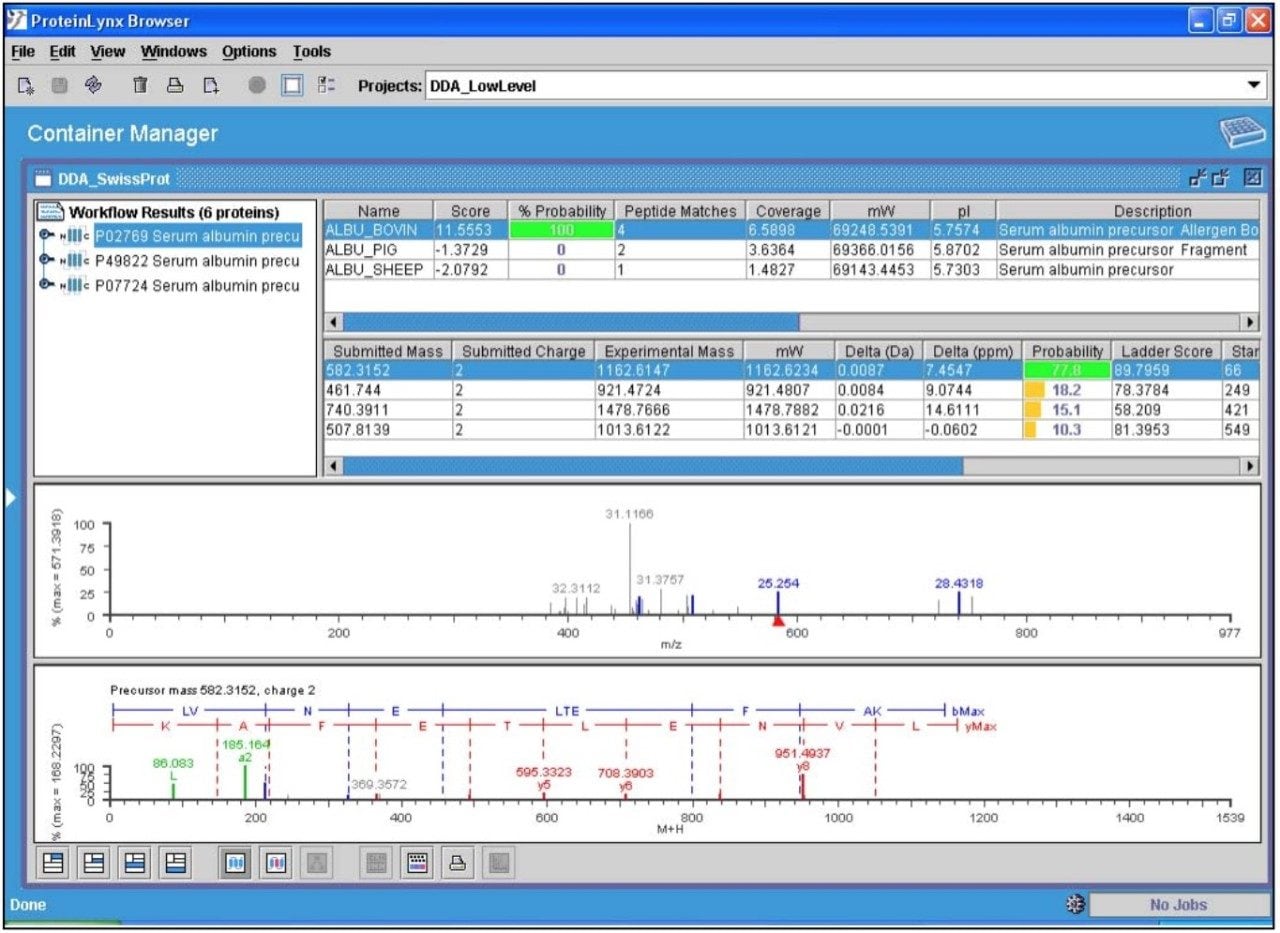
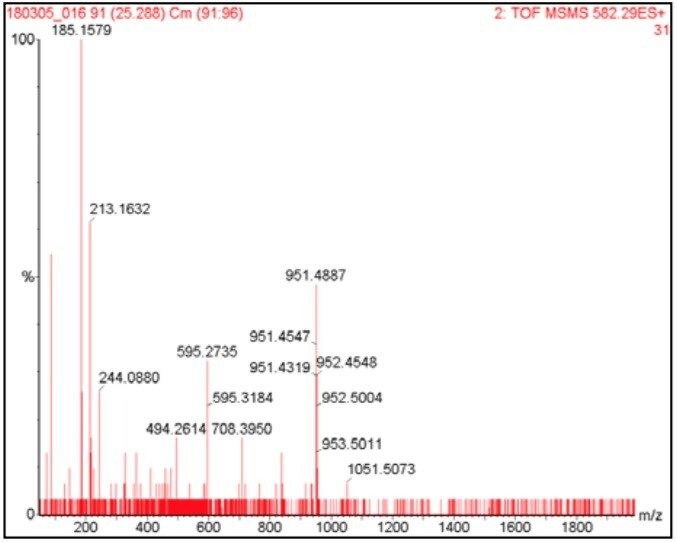
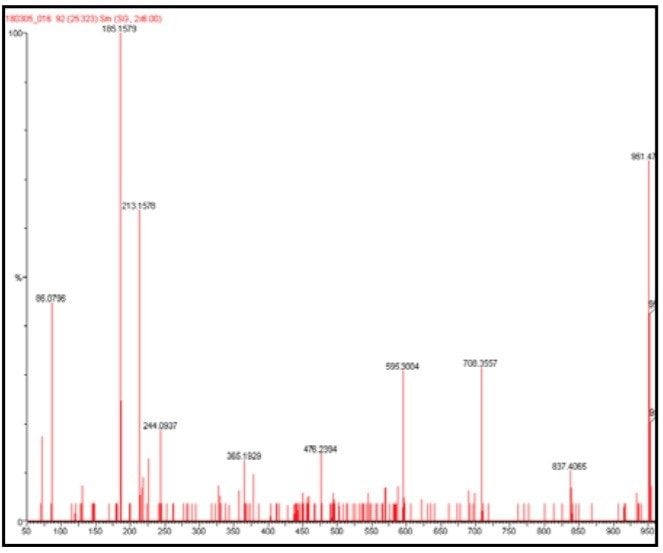
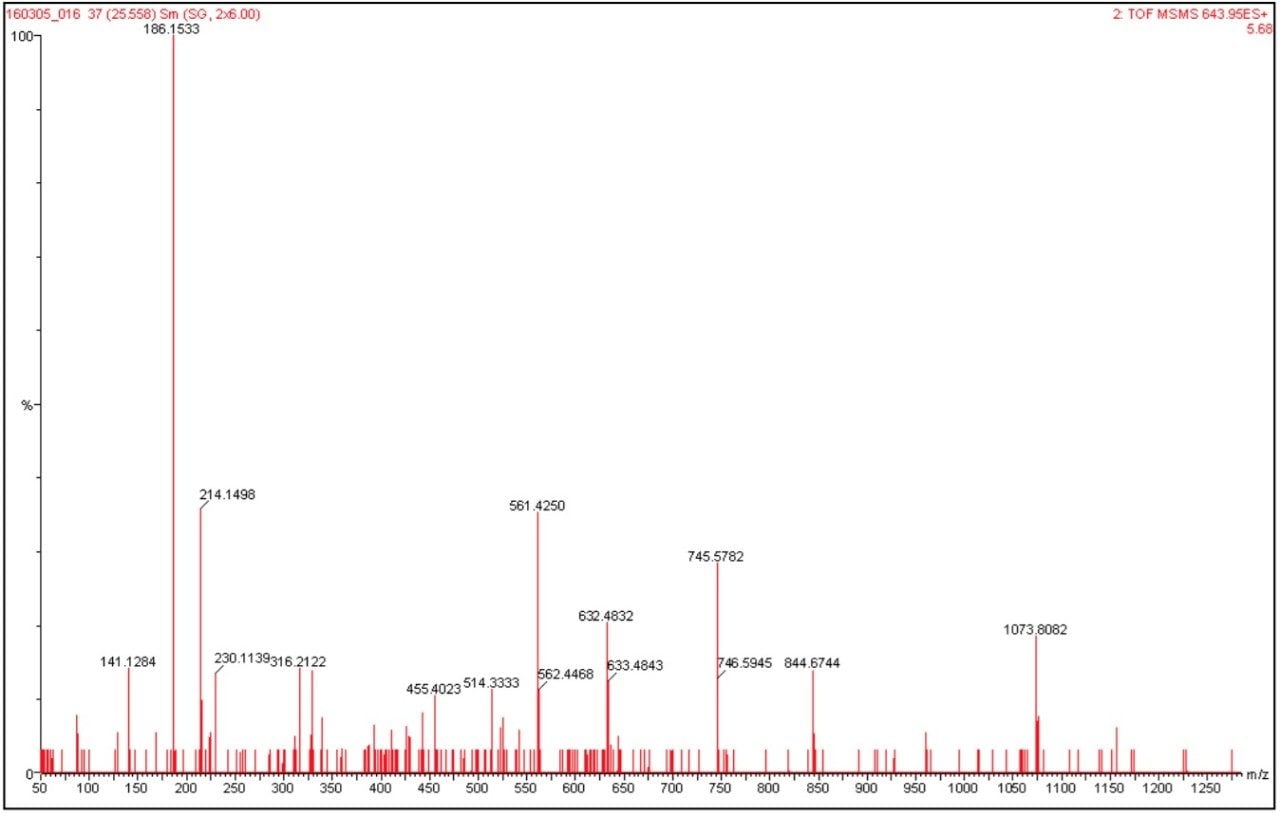
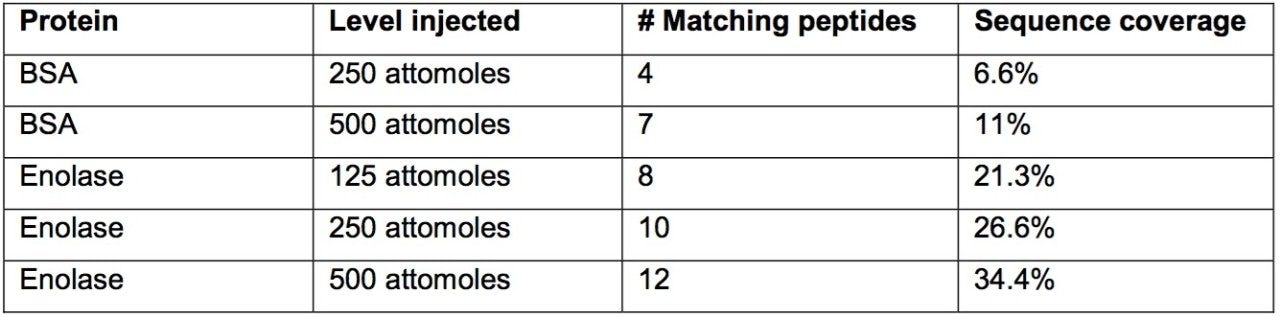
The data presented below illustrates typical results that can be obtained from low-level tryptic digests using the nanoACQUITY/Q-Tof Premier combination in DDA mode. Example data from two digest samples, Bovine Serum Albumin (BSA) and Yeast Enolase are shown. In the case of BSA, data is presented from 250 and 500 attomoles, while in the case of Enolase, data is shown from 125, 250 and 500 attomoles. In each case positive identification of the parent protein could be made and is shown using ProteinLynx Global SERVER v2.2.
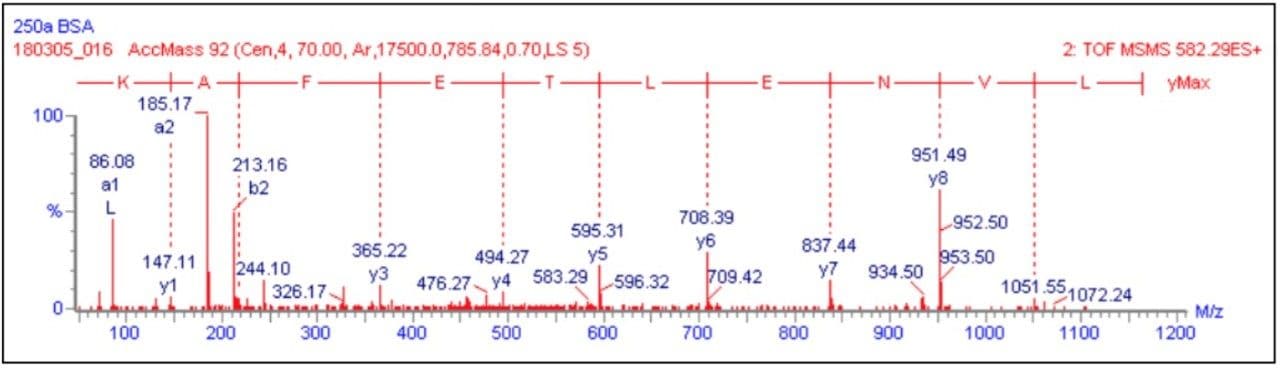
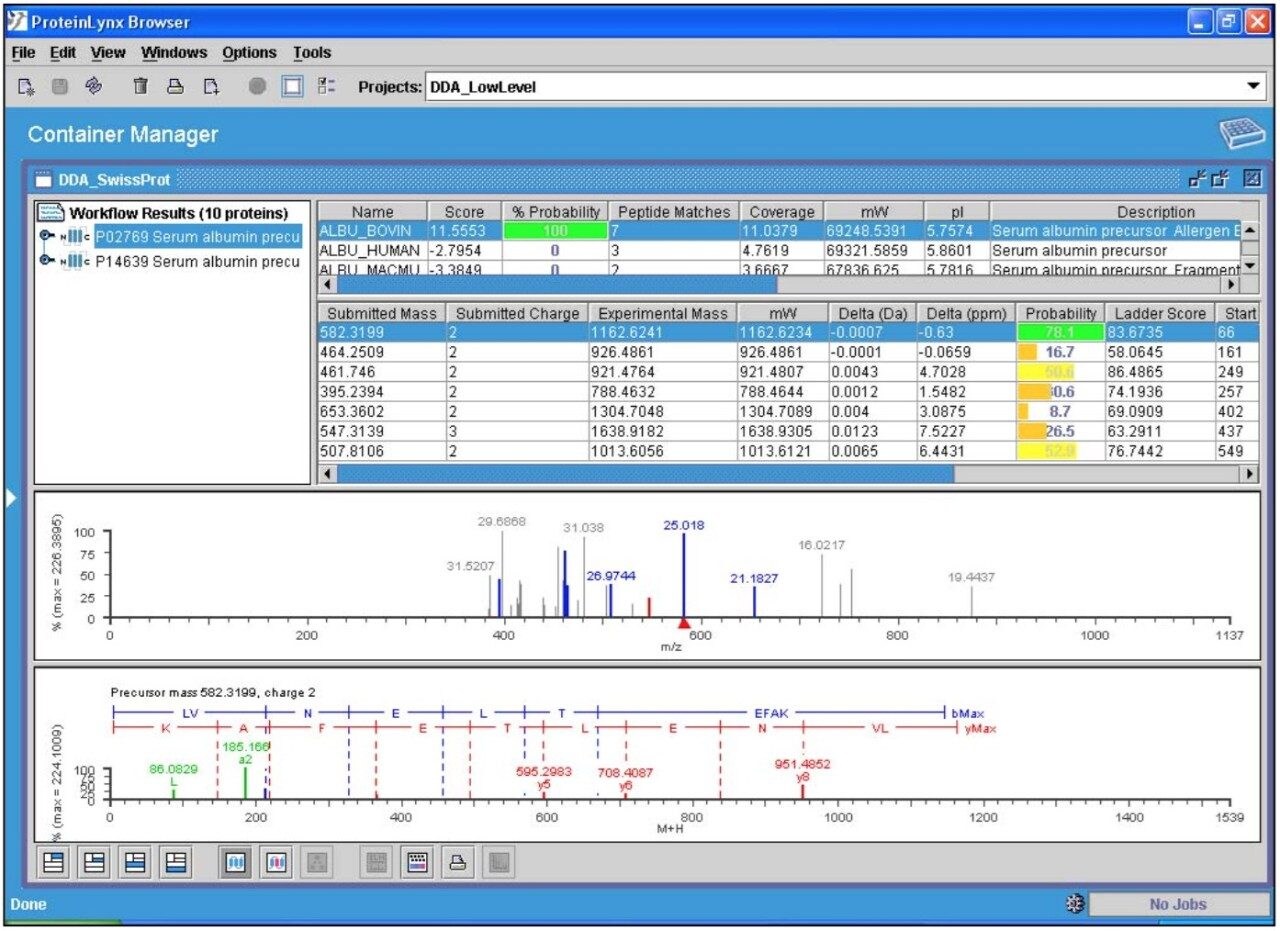
720001252, May 2005