For research use only. Not for use in diagnostic procedures.
This is an Application Brief and does not contain a detailed Experimental section.
This application brief demonstrates the improvements in Waters High Definition Imaging (HDI) Software, which now provides a more intuitive user interface and streamlined acquisition process, including seamless integration of instrument control MassLynx Software. With these improvements, multiple experiments can be queued and the data can be processed automatically for review within HDI.
New software acquisition approach streamlines DESI imaging workflow to maximize data acquisition.
DESI (Desorption Electrospray Ionization) mass spectrometry is a simple and straightforward mass spectrometry imaging technique. It requires no sample preparation, is performed at ambient pressure, and provides good sensitivity for a range of compounds. Recent hardware improvements have made DESI a robust and stable technique, allowing DESI imaging experiments to be run at a spatial resolution in the region of 50 µm. Now the demand is on the MS acquisition system to ensure that maximum information can be obtained from each tissue sample. This requires fully automated sequential experiments – for example, the acquisition of multiple DESI imaging datasets from samples mounted on the same or adjacent glass slides. Without an integrated software solution however, sequential DESI imaging experiments can be challenging to set up, with multiple parameters to control.
An improved version of Waters High Definition Imaging Software – combined with the implementation of DESI imaging in MassLynx – has been developed to provide a single, streamlined, and user-friendly acquisition process.
This new DESI imaging workflow (Figure 1) consists of:
Once these simple steps are complete, the whole experiment is exported as a MassLynx sample list to be run directly on the system without any further input. MassLynx controls the stage of the Prosolia 2D DESI source during the imaging experiment – including the X/Y coordinates – acquiring all raw data into a single data file per image.
Following acquisition of the DESI imaging data, the raw data can be processed automatically (Figure 1e) such that it can be visualized in the Image Control section of HDI (Figure 1f), including the full integration of ion mobility separation, if present in the data. The DESI ion images and the optical image can also be easily overlaid.
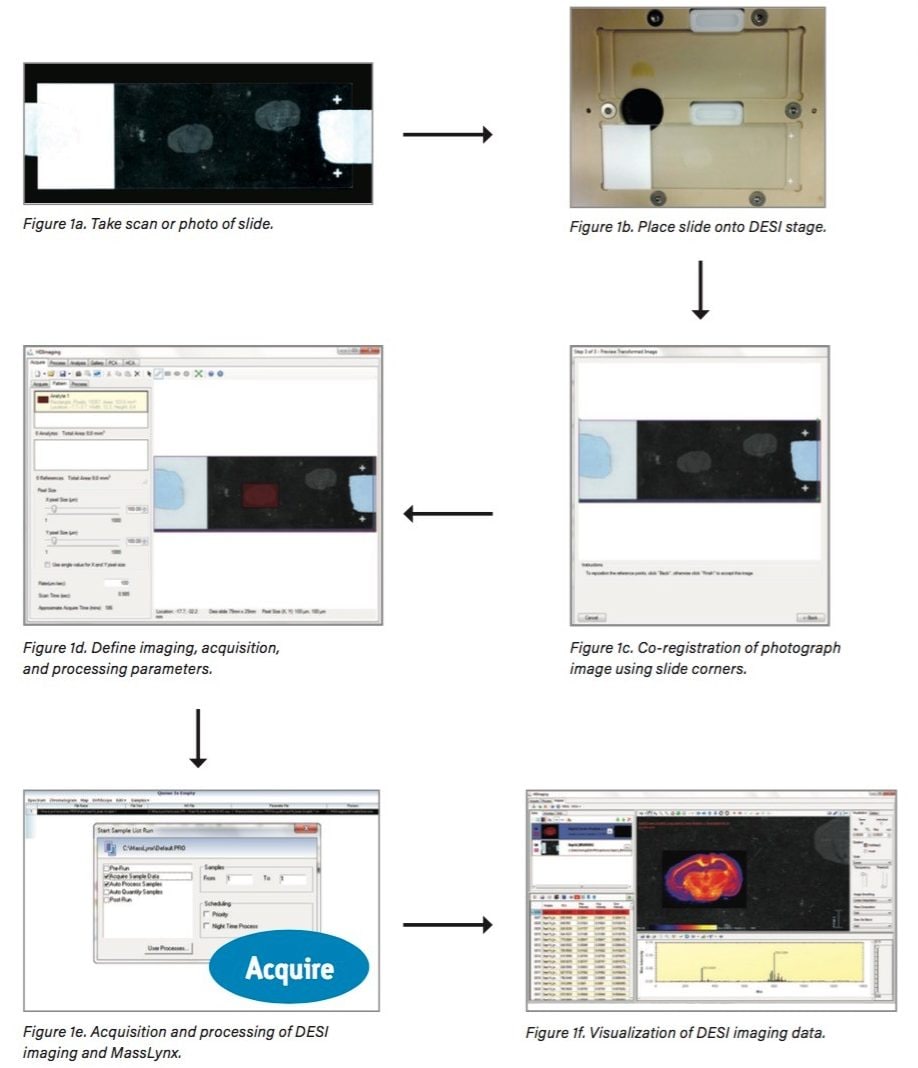
An additional advantage of this experimental approach is that multiple experiments can be queued up to run within MassLynx. Therefore, a selection of samples can be analyzed without the need for user intervention.
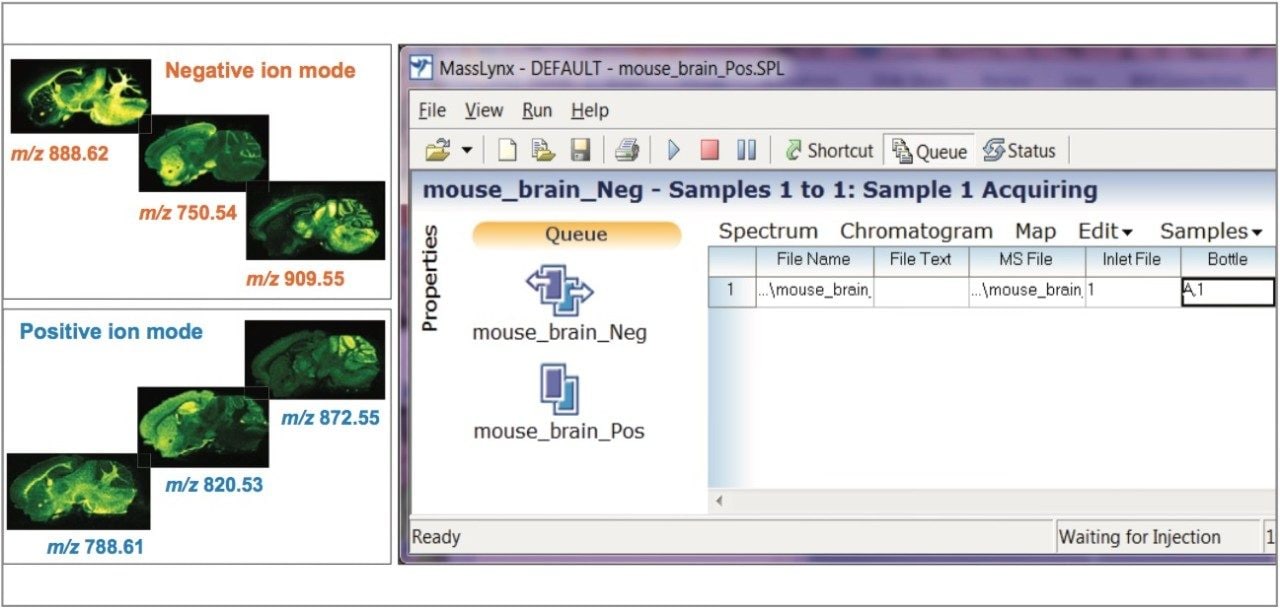
In Figure 2, this approach has been used to analyze a mouse brain tissue section, first in negative ion mode, followed directly by positive ion mode of acquisition over the same area of the tissue. At the end of both experiments, the two datasets were automatically processed and ready to be reviewed by the user within HDI.
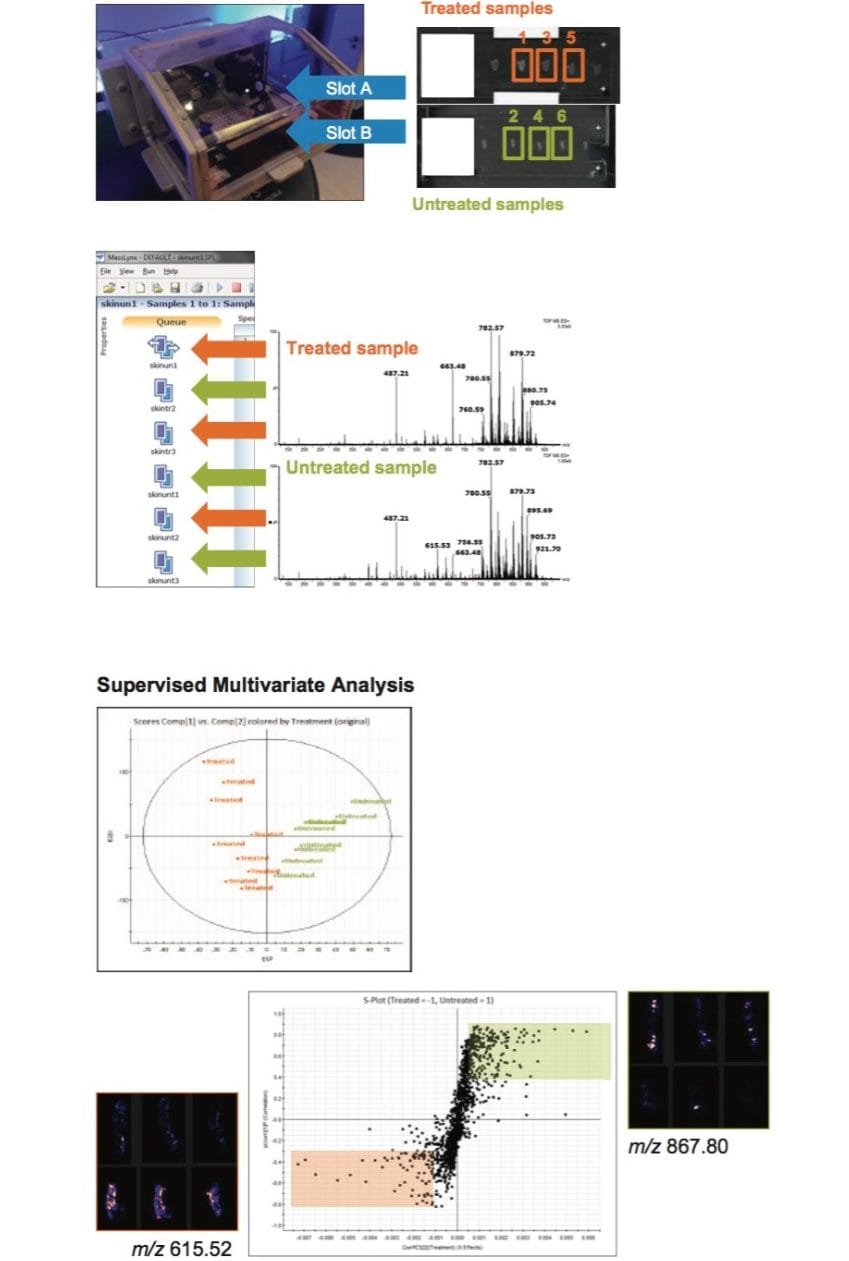
Another example is displayed in Figure 3, showing the workflow used on three samples of drug-treated mouse skin tissue sections placed onto one slide, and three samples of tissue sections from an untreated mouse placed onto a second slide. By mounting these slides into the two holders of the DESI source, six imaging experiments were run in sequential fashion, eliminating the need for user intervention and allowing a complete imaging MS study to be run overnight. With automated processing of the six raw datasets, it was possible to carry out unsupervised and supervised multivariate analyses – Principal Component Analysis (PCA) and Orthogonal Partial least squares Discriminant Analysis (OPLS-DA) – to extract the molecular differences between the treated and untreated skin tissue sections.
We thank Prof. Anna Nicolaou and Dr. Alexandra Kendall from Manchester Pharmacy School for providing the samples for the multiple DESI imaging experiment.
720005651, April 2017