
In this application note, foodstuffs of plant origin were further cleaned using Oasis PRiME HLB following QuEChERS extraction and run on the Xevo TQ-GC to quantify 208 pesticides and their metabolites in fruits and vegetables. This simple pass-through cleanup is readily incorporated into the QuEChERS workflow to maintain accuracy and precision in the quantitative performance, while improving overall method robustness. Rigorous method verification was carried out following the SANTE/11813/2017 guidance document, which provided strong evidence that the method is fit for purpose to achieve the Chinese National Standard Method regulatory requirements for GC-MS/MS pesticides (GB 23200.113-2018).
Efficient workflows enable reliable determination of multiple residues across a variety of challenging food commodities. Waters offers a range of sample preparation techniques that provide improved accuracy for quantifying contaminants.
Gas chromatography-mass spectrometry (GC-MS) has been a common analytical method for pesticide measurement due to its high efficiency of separation, along with its qualitative and quantitative performance. As a milestone of pesticides analysis, Lehotay1 and Nguyen, et al.2 established a sample preparation method based on QuEChERS technology in 2015 for the simultaneous detection of multiple pesticide residues in vegetables and other foods using LC-MS/MS and GC-MS/MS. In recent years GC-MS/MS analysis has become the preferred method for pesticides analysis due to its advantages in selectivity, sensitivity, high throughput, and accurate quantitative performance.3
Recently, the first Chinese National Standard Method (GB 23200.113-2018)4 for multiple pesticide residues using GC-MS/MS was released. For the first time in GB methodology, two efficient technologies have been adopted: QuEChERS for sample extraction, and GC-MS/MS for detection.
In this application note, foodstuffs of plant origin were further cleaned using Waters Oasis PRiME HLB following QuEChERS extraction and run on the Xevo TQ-GC to quantify 208 pesticides and their metabolites in fruits and vegetables. Rigorous method verification was carried out following the SANTE/11813/2017 guidance document,5 which provided strong evidence that the method is fit for purpose and will achieve the method validation criteria set by the GB 23200.113-2018.
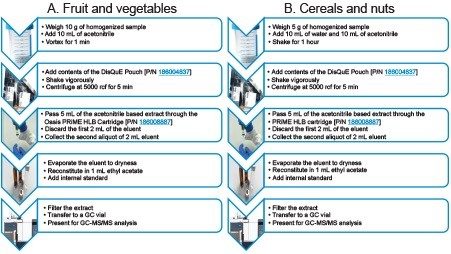
Cucumber, grape, and rice samples were purchased from local retail outlets and prepared using a modified version of QuEChERS sample preparation as reported in CEN method 15662.6 The sample preparation used is summarized in Figure 1.
|
Column: |
Rtx-1701 (30 m x 0.25 mm x 0.25 μm) |
|
Carrier gas: |
Helium |
|
Gas flow rate: |
1.0 mL/min |
|
Injection type: |
Pulsed splitless |
|
Injection liner: |
Gooseneck splitless 4 mm x 6.5 x 78.5 (Restek) |
|
Inlet temp: |
280 °C |
|
Pulse time: |
1.0 min |
|
Pulse pressure: |
170 kPa |
|
Purge flow: |
30 mL/min |
|
Septum purge flow: |
3 mL/min |
|
Wash solvent: |
Hexane |
|
Oven program: |
80 °C (hold 1.1 min) to 120 °C at 40 °C/min, then to 240 °C at 5 °C/min, then 295 °C at 12 °C/min and hold 8 min |
|
Run time: |
38.68 min |
|
Injection volum: |
1 μL |
|
MS system: |
Xevo TQ-GC |
|
Software: |
MassLynx v4.2 |
|
Ionization mode: |
EI, 70 eV |
|
Source temp.: |
250 °C |
|
GC interface: |
300 °C |
|
MRM conditions: |
All transitions were imported from the Waters Quanpedia Database. IntelliStart Custom Resolution settings were used. |
Typically for GC, pigments are undesirable because they can potentially contaminate the injection liner and the GC column. Graphitized carbon black (GCB) is commonly used to remove pigments. However caution is advised with the level of GCB used since it is both a reverse phase and an anion exchange sorbent and can potentially trap certain pesticides, especially for pesticides with planar structure. Therefore it is important to optimize the amount of GCB used to capture the maximum amount of pigment while maintaining good recovery of pesticides, which can be a time-consuming exercise. In this work GCB was not used, but instead a novel sorbent, Oasis PRiME HLB was employed. Oasis PRiME HLB has recently been used to quickly and efficiently remove co-extractives including fats and phospholipids, as well as pigments from food matrices, using a simple and fast pass-through protocol.7 In this study, Oasis PRiME HLB provided excellent pigment removal, thus reducing the contamination of the GC inlet liner and extending the lifetime of the GC consumables.
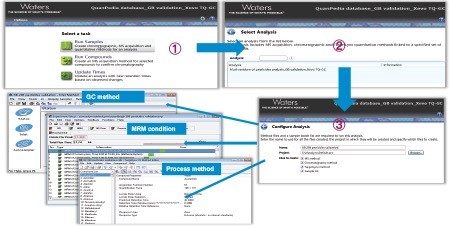
GC-MS/MS methods for GB 23300.113-2018 were easily generated using Quanpedia Database. This provided the creation of the GC, MS/MS, and processing methods in three simple clicks, as shown in Figure 2. Quanpedia can greatly reduce time and lab resources employed for setting up new multi-residue methods.8
In-house method verification was carried out to determine the overall method performance in accordance with the requirements of GB method 23300.113-2018, referencing the SANTE/11813/2017 guidance document and associated analytical and validation criteria.5 The method performance was assessed for trueness, reproducibility, quantification, and identification of 208 pesticides and associated metabolites in cucumber, grape, and rice. For each commodity (n=3), matrix matched calibration curves were generated and replicate spikes (n=6) were extracted at three concentrations (LOQ, 2x LOQ, and 5x LOQ. The results, as summarized in Table 1, were within the permitted tolerances of the required guidelines demonstrating that this method is fit for purpose.
|
Parameter |
SANTE criteria |
Rice |
Grape |
Cucumber |
Criteria satisfied |
|---|---|---|---|---|---|
|
Retention time |
±0.1 minute |
20.49–20.50 |
18.69–18.70 |
18.67–18.70 |
Yes |
|
Ion ratio |
±30% |
1.92–2.28 |
1.55–2.43 |
1.92–2.28 |
Yes |
|
Residuals |
±20% |
≤20% |
≤20% |
≤20% |
Yes |
|
Recovery (trueness) |
70 to 120% |
103.6% |
93.4% |
96.9% |
Yes |
|
Repeatability (RSDr) |
≤20% |
2.6% |
3.5% |
2.8% |
Yes |
|
LOQ |
≤MRL |
0.02 mg/kg |
0.01 mg/kg |
0.01 mg/kg |
Yes |
Table 1. Summary of the in-house verification results for pesticides and associated residues in rice, cucumber, and grape at relevant concentrations (LOQ, 2x LOQ, and 5x LOQ).
Trueness and repeatability were assessed from the analysis of the three commodities: cucumber, grape, and rice. Each commodity was spiked at three concentration levels: LOQ, 2x LOQ, and 5x LOQ with five replicates (n=5) of each concentration prepared.
In this study, the method performance is reported for each commodity spiked at the LOQ only, namely cucumber at 0.01 mg/kg, grape at 0.01 mg/kg, and rice at 0.02 mg/kg. These spiked concentrations were selected based on the LOQs defined in GB method 23200.113-2018. Figure 3 shows the chromatograms from some of the pesticides spiked at 0.01 mg/kg in rice, demonstrating that the sensitivity for these compounds is much lower than the required LOQ specified in the GB method.
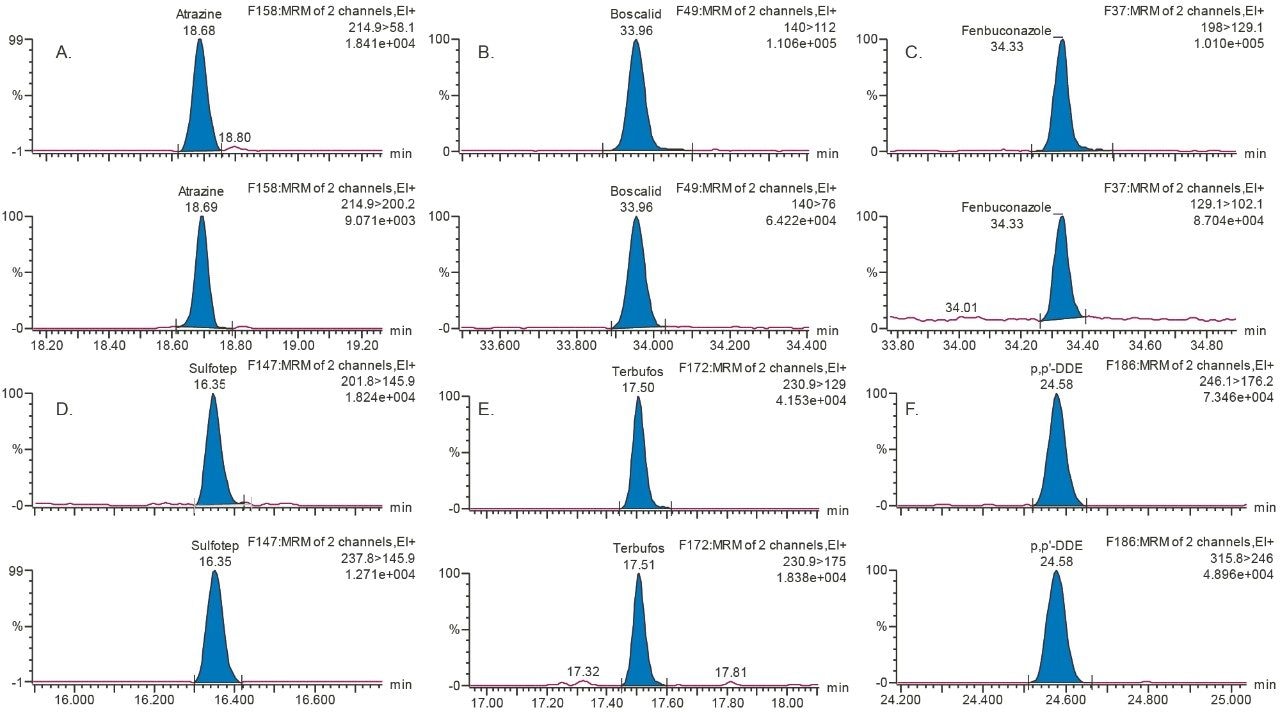
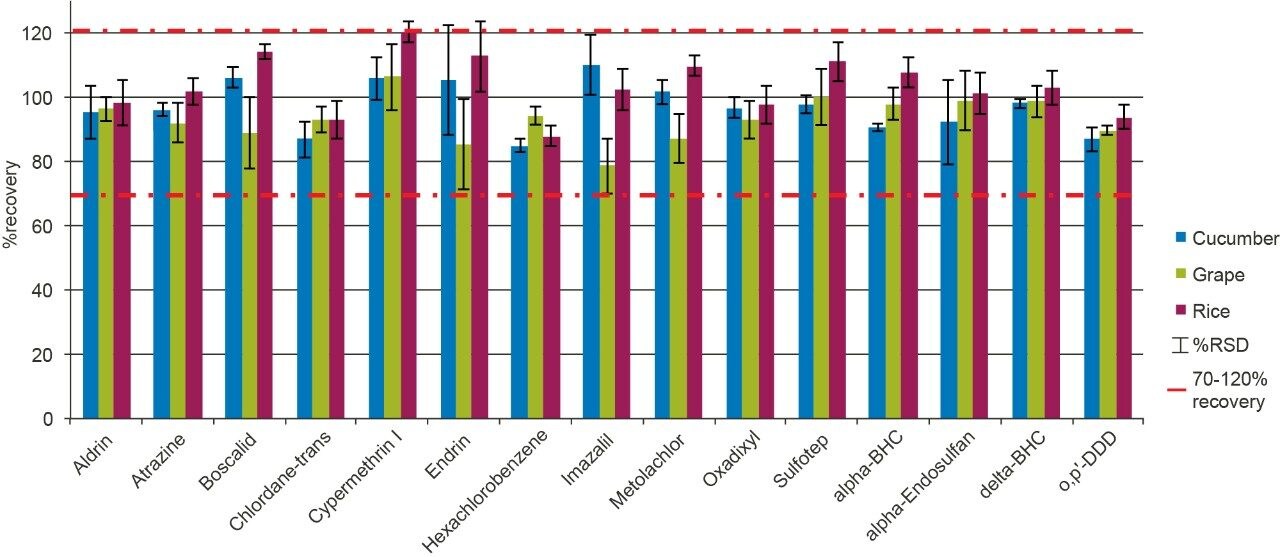
Figure 4 shows the measured percentage recovery (trueness; between 70 and 120%) and repeatability (%RSD; <20%) for a representative selection of 15 pesticides in all of the commodities tested. Further details on recovery and repeatability for all 208 pesticides at the required LOQ across each commodity are summarized in Table 2, in the Appendix, which meet the acceptance criteria of the GB method.
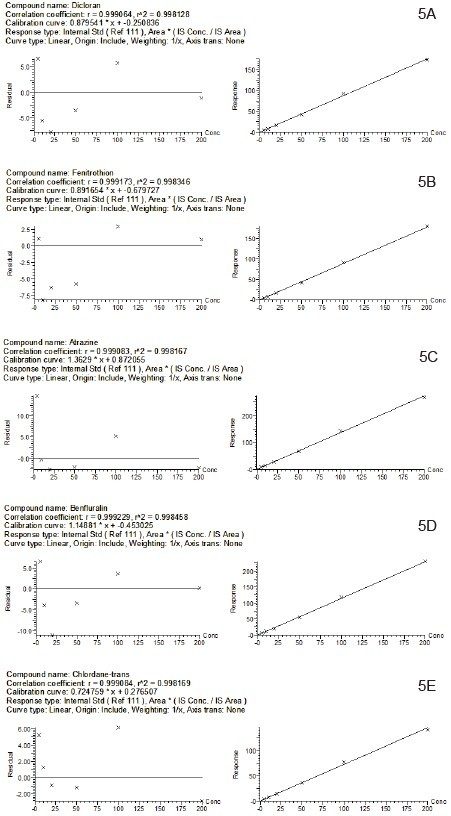
Matrix-matched calibration curves allowed for accurate quantification of pesticides spiked in the commodity at the required LOQs. Calibrations were prepared over the concentration range of 0.005 mg/kg to 0.2 mg/kg for each target compound using internal standards. A weighted linear regression (1/x) was applied. Individual back-calculated concentrations were calculated automatically by TargetLynx Application Manager, and all were within the tolerance set in the SANTE guidelines (±20%). Figure 5 shows matrix-matched calibration plots for five representative pesticides.
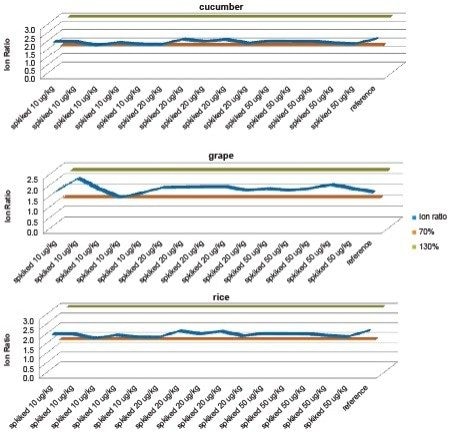
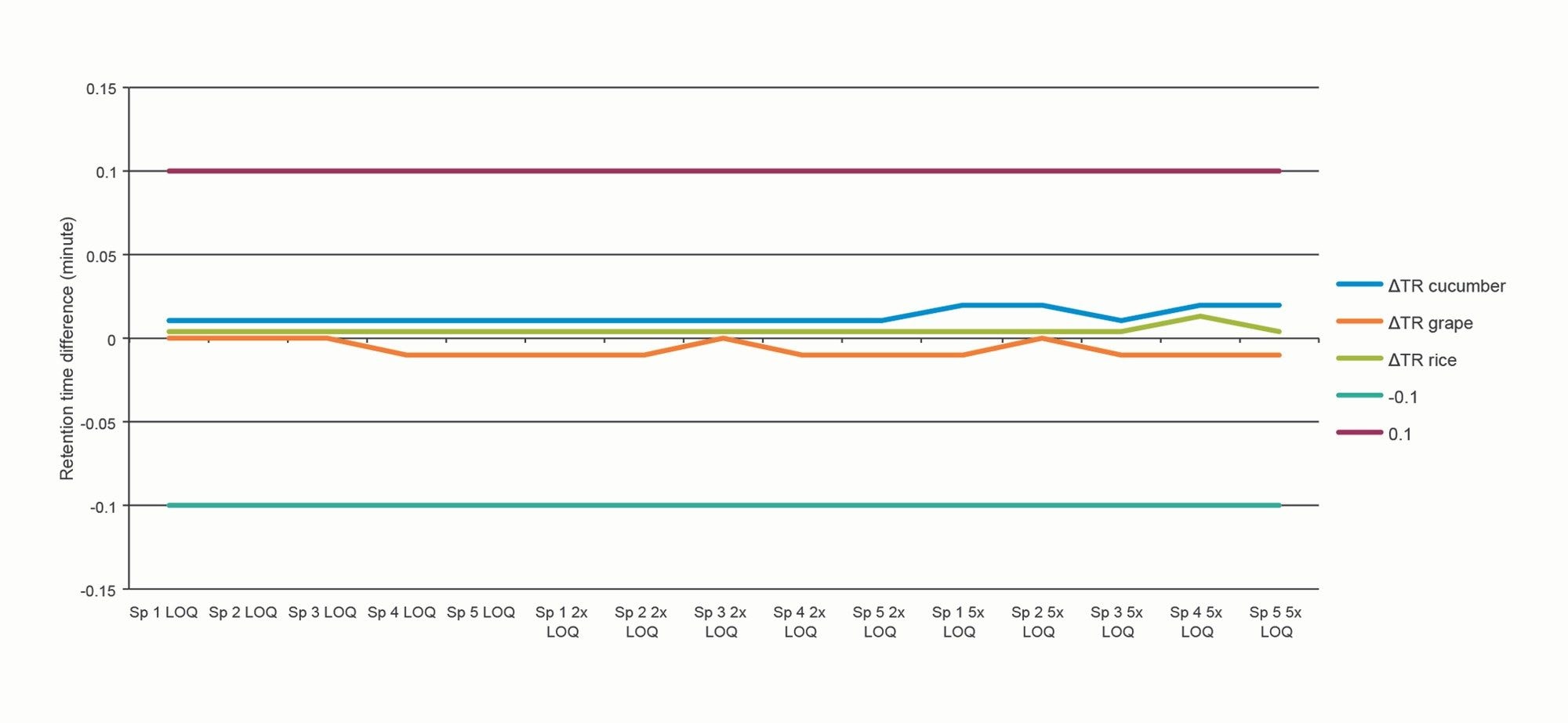
The GC-MS GB Methods reference the SANTE requirements with respect to retention time and ion ratio tolerances. The guidelines state that the retention time of the analyte in the extract should be ±0.1 min to that of the calibration standard, and that ion ratios from sample extracts should be within ±30% of the reference (averaged calibration standards in the same sequence).
Using atrazine as an example, Figure 6 and Figure 7 show the plot of ion ratios and delta retention time, demonstrating that the analytical criteria within the guidelines were met.
720006376, October 2018