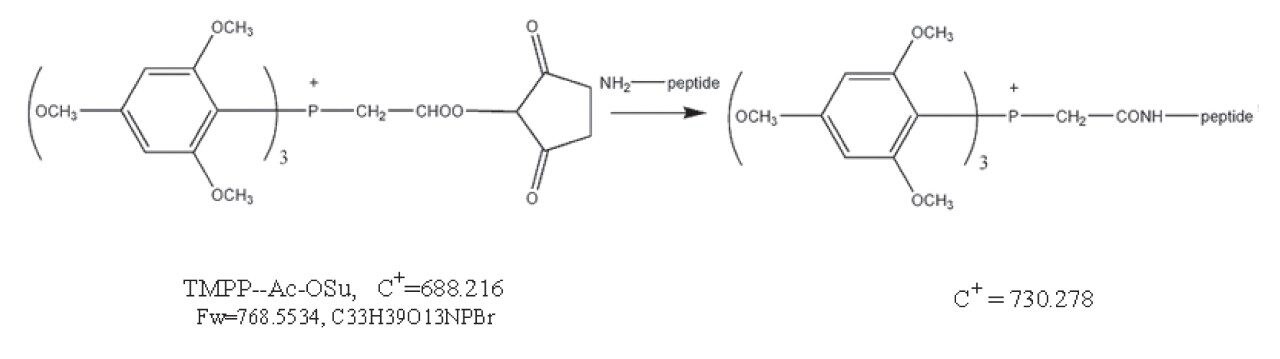
This application note describes the use of an N-terminal derivatization strategy, using the N-Tris (2,4,6-trimethoxyphenyl) phosphonium-acetic acid N-hydroxysuccinimide ester (TMPP-acOSu) to improve MALDI MS/MS fragmentation patterns. Coupling of this peptide derivatization protocol with nanoscale HPLC separation and deposition onto a MALDI target is presented.
Analysis of TMPP derivatized tryptic peptides in the MS mode can improve the amino acid sequence coverage of the proteins identified in a PMF experiment
Peptide mass fingerprinting (PMF) by MALDI Tof mass spectrometry is a rapid and sensitive method for the identification of proteins from organisms with well characterized genomes. PMF compares experimentally observed peptide masses, obtained from the MALDI MS of enzymatically digested proteins, with theoretical peptide masses, obtained from the in silico digestion of proteins contained within protein sequence databanks. This is a highly specific, sensitive technique for identifying proteins from a known proteome. However, the PMF approach will fail when a protein is not represented in the protein sequence database (e.g. if the genome has not been sequenced). In addition, problems occur due to non-specific enzyme activity, if some of the peptides are modified or too few tryptic peptides are observed to give an unambiguous answer. In these cases, further analysis and added specificity is gained by performing MS/MS on some of the peptide molecular ions and searching this information against a sequence database. If this fails to give an unambiguous answer then homology-based searching of the amino acid sequence information may allow identification of the parent protein.
With a hybrid quadrupole orthogonal acceleration time-of-flight (Q-Tof) mass spectrometer, equipped with a MALDI source, high quality MS/MS spectra can quickly be generated from MALDI phase samples. A feature of the oa-Tof mass analyzer is the high mass accuracy that is routinely achieved—typically better than 10 ppm RMS when using a single-point lockmass. Despite the quality of the MS/MS data obtained from this geometry of instrument, the MS/MS spectra generated from singly charged peptides, produced by the MALDI ionization process, exhibit a wide variety of fragment ions. The types of fragment ions produced are very sequence dependent. This can make interpretation of MALDI MS/MS data difficult due to their complexity.
A way of simplifying the spectrum and improving the fragmentation efficiency is to drive the fragmentation process by introducing a fixed charge at a specific location on the peptide.
This application note describes the use of an N-terminal derivatization strategy, using the N-Tris (2,4,6-trimethoxyphenyl) phosphonium-acetic acid N-hydroxysuccinimide ester (TMPP-acOSu) to improve MALDI MS/MS fragmentation patterns. Coupling of this peptide derivatization protocol with nanoscale HPLC separation and deposition onto a MALDI target is presented.
One advantage of this derivatization reagent is, that it increases the peptide masses by 572.182 Da. This mass increase can bring small peptides into the m/z range 800–3500, the typical range used in PMF analysis. A theoretical tryptic digestion of 100 randomly selected proteins from the Swiss-Prot databank revealed that small tryptic peptides, with fewer than eight amino acid residues, account for ca 20% of protein coverage.

A lyophilized sample of an ADH digest (Waters, Milford, MA) was dissolved in 100 μL of triethylammonium bicarbonate (Sigma, St Louis, MO)/Acetonitrile (80/20), at a concentration of 10 pmoles/μL. The peptide solution was made up in Eppendorf tubes (Eppendorf, Germany).
2.5 mg of TMPP-acOSu (Waters, Milford, MA) was dissolved in 60 μL anhydrous acetonitrile, and 0.25 μL of this solution was added to 25 μL of the tryptic digest. The mixture was vortexed and left tostand for 20 minutes. The reaction was subsequently acidified using TFA, and the mixture combined with 5 mg/mL of MALDI matrix solution (alpha-cyano-4-hydroxycinnamic acid). 1 μL of the resulting solution was spotted onto a stainless steel MALDI target plate (Waters, Manchester, UK).
All data were acquired on a Waters Micromass Q-Tof Ultima MALDI Mass Spectrometer in positive ion mode. The instrument was calibrated with a mixture of PEG 200, 600, 1000, and 2000. In MS mode, data were acquired from m/z 800 to 3000. A lockmass, (Glu1)-Fibrinopeptide B, from an adjacent well was used as a near point calibration reference, to enhance the mass measurement accuracy of the data. In the MS/MS mode, the data were acquired from 50 Da to an m/z value of 5% above the parent ion [M+H]+.
The analytical system consisted of a Waters CapLC XE System configured with a ten-port switching valve, the stream select module. The HPLC was configured with a Waters Symmetry C18 3.5 μm, 320 μM x 150 mm Column. The flow rate was 3 μL/min and peptides were separated using the following solvents: Water + 0.1% TFA and acetonitrile + 0.1% TFA. The initial solvent composition of 97% water (with 0.1% TFA)/3% acetonitrile was held for the first 3 minutes. A gradient was run with the organic solvent rising up to 60% in 27 minutes, and then to 95% by 30 minutes. The acetonitrile composition then stayed at 95% for 3 minutes to wash the column. Re-equilibration of the column was achieved using the initial conditions for five minutes.
The outlet from the capillary scale HPLC column was attached directly to the MALDI spotting robot, the Waters 2700 MS. The eluent from the HPLC column was spotted onto the MALDI target, with each spot corresponding to 20 seconds of chromatographic elution.
Waters ProteinLynx Global SERVER v2.0 was used for data processing and database searching. Data were converted to XML format and searched against the Swiss-Prot database v40. MS/MS spectra were deisotoped using MaxEnt 3 (MaxEnt Solution, UK). De novo sequencing was performed using Waters MassSeq Software.
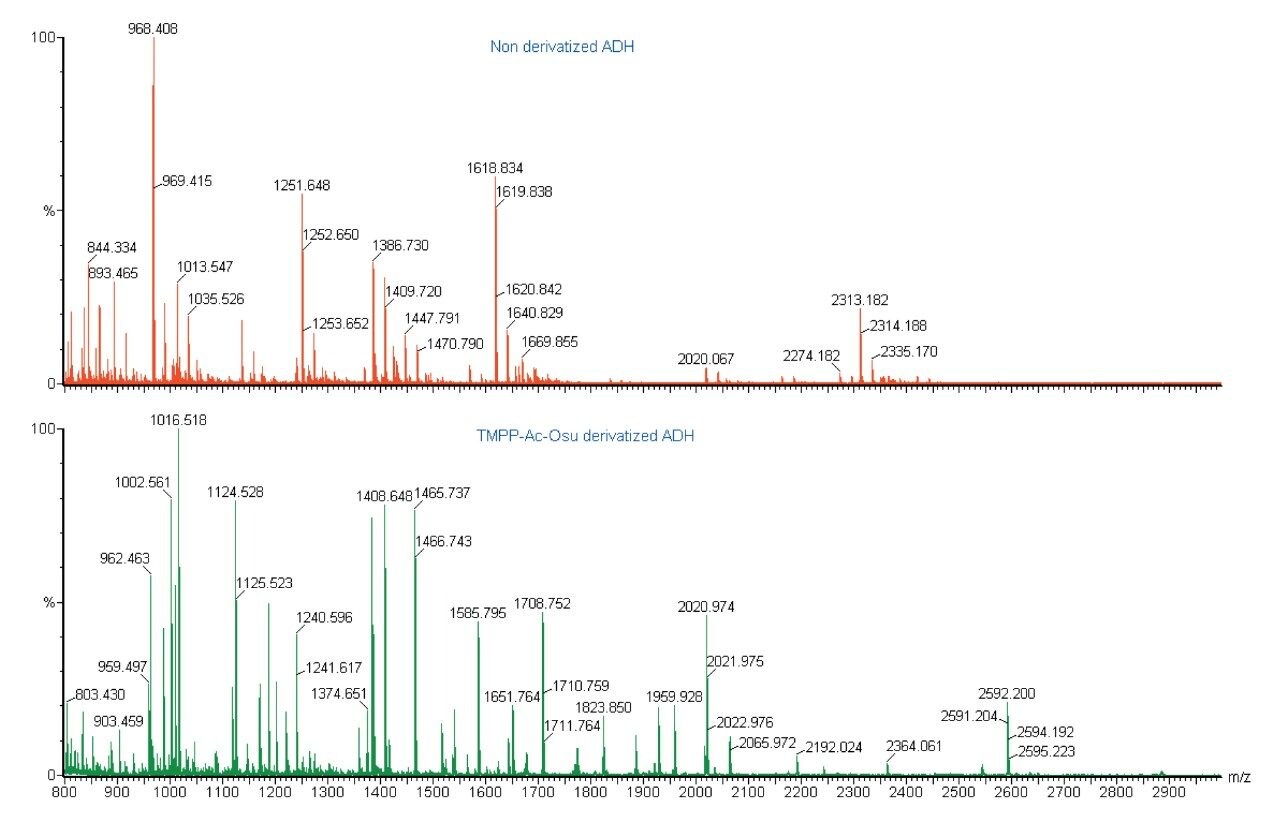
Analysis of a native (non-derivatized) ADH tryptic digest yields an average of 10 peptides in a MALDI mass spectrum. This equates to an amino acid sequence coverage of approximately 43% of the protein sequence.
In contrast, analysis of the ADH digest after derivatization with TMPP, resulted in 40 peptides being observed in the mass spectrum. MS/MS experiments were subsequently performed on 35 of these using the Q-Tof Ultima MALDI Mass Spectrometer.
Of the peptides identified in the modified mass spectrum that were not seen in the unmodified spectrum, 15 had a mass below 850 Da (which is the lower mass limit in MALDI MS due to the interference of the matrix related peaks). This illustrates the significant number of small peptides, which are present, but not observed in a typical tryptic digest. However, these are observed, following TMPP derivatization of the tryptic peptides. The percentage amino acid sequence coverage, in this case, was increased from 43% to 62% by derivatizing with TMPP-oSu.
The same TMPP-derivatized ADH digest sample has been analyzed by an LC-MALDI approach. In this case the number of peptides analyzed is even greater, with 120 peptides detected, of which 105 were identified using MS/MS on the MALDI Q-Tof. Due to the chromatographic separation of the peptides present, the number of peptides identified was significantly improved, resulting in a rise in the sequence coverage to 84%. The combination of HPLC separation with MALDI MS detection dramatically increased the dynamic range of the experiment.
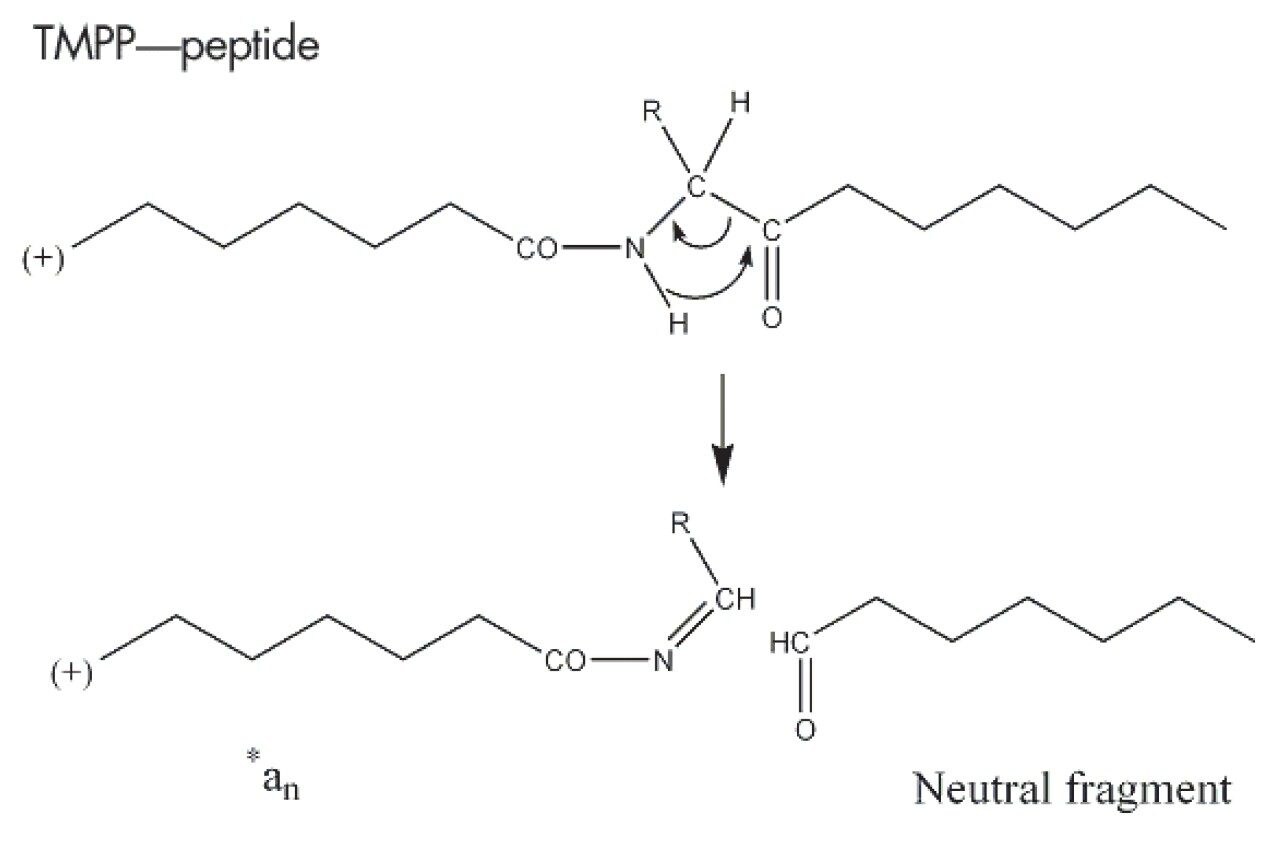
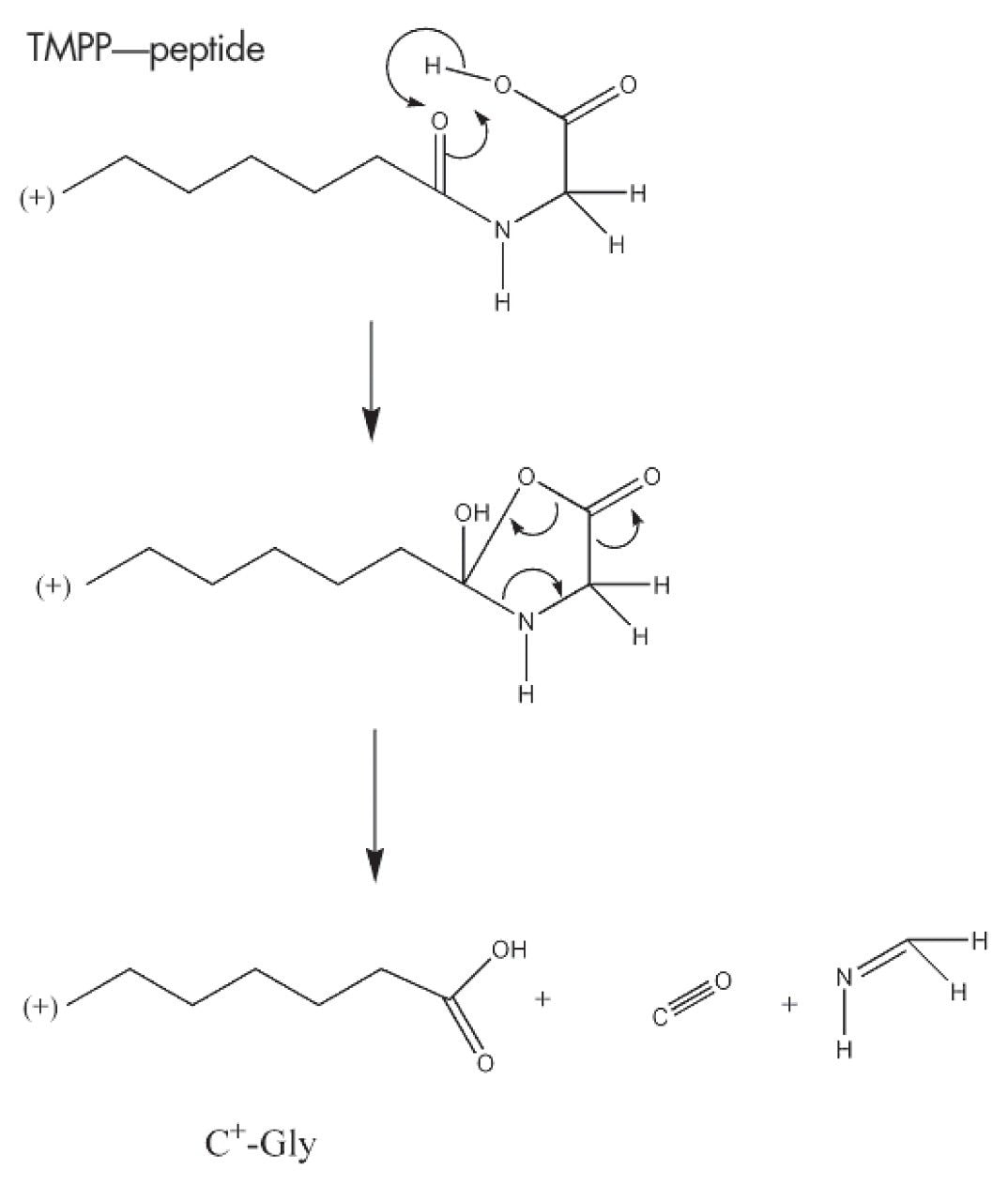
The fragmentation patterns observed in MS/MS for TMPP-derivatized peptides are more predictable than their non-derivatized peptide counterparts. This has previously been studied by several groups, with the most extensive study performed by Sadagopan et. al.1 In all of the MS/MS spectra obtained from TMPP modified peptides, fragment ions are observed between 573.1898 Da and the precursor ion mass. The 573.1898 Da ion is the M+ of the TMPP reagent. This ion at m/z 573.1898 is typically a strong peak in the spectrum, making it a diagnostically useful species, allowing confirmation that the analyzed precursor ion has been derivatized with TMPP. Fragment ions present below m/z 573.1898 are predominantly due to fragments produced from cleavage of the TMPP molecule. Even if *an or *bn type ion peaks are not intense compared to precursor ion or to the ion at 573.1898 Da, it is possible to detect and identify them. As internal rearrangement is not favored from these derivatized species, noise in the MS/MS spectrum obtained from TMPP derivatized peptide molecules is very low.
After analyzing a significant number of TMPP derivatized MS/MS spectra from the ADH tryptic digest, it can be concluded that:
The following spectra are illustrations of the differences in fragmentation between underivatized (bottom spectra) and derivatized (top spectra) ADH peptides on the Q-Tof Ultima MALDI.
Complete sequence was obtained for both fragmentations. However the derivatized peptide gave less re-arrangement fragmentation peaks, which make the interpretation easier.
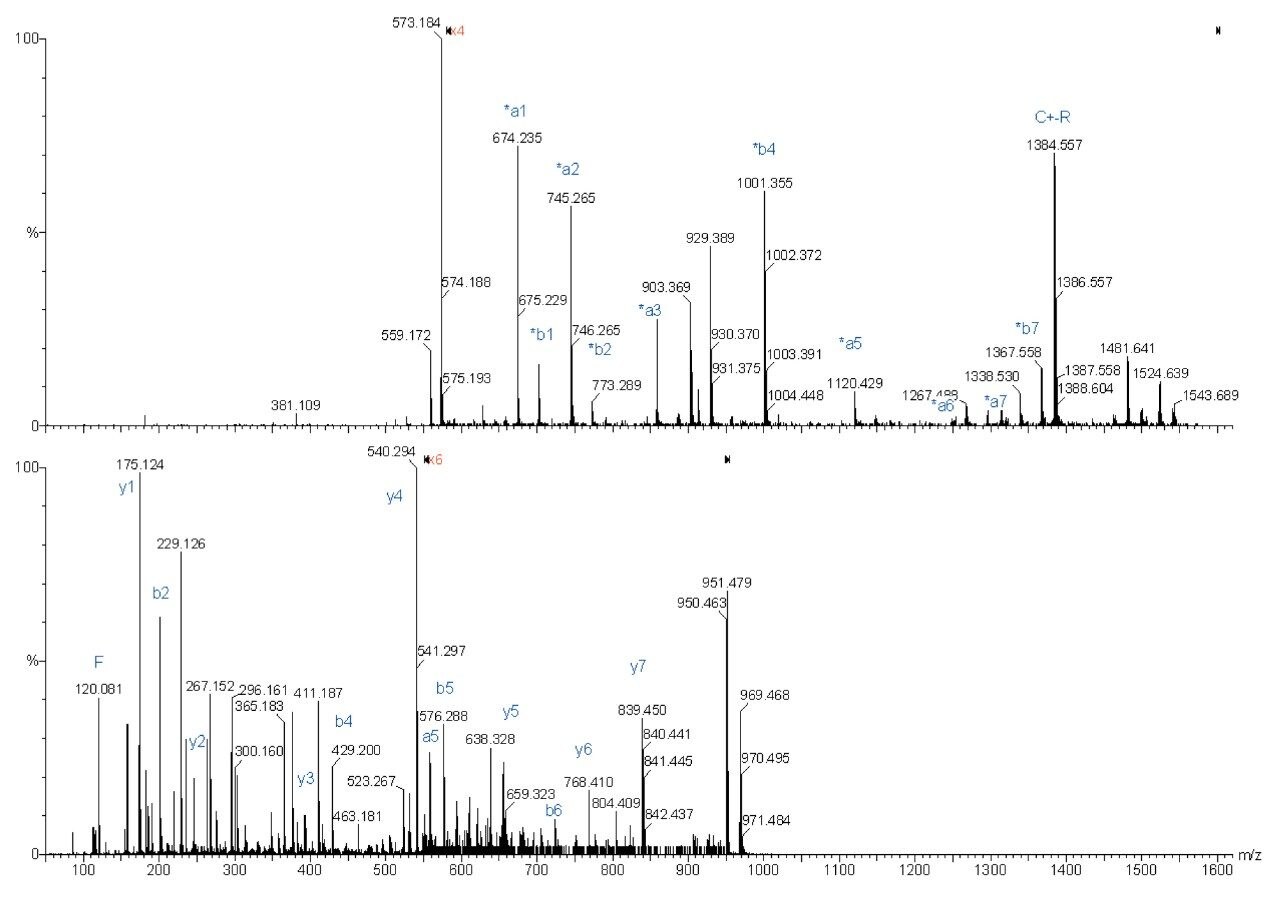
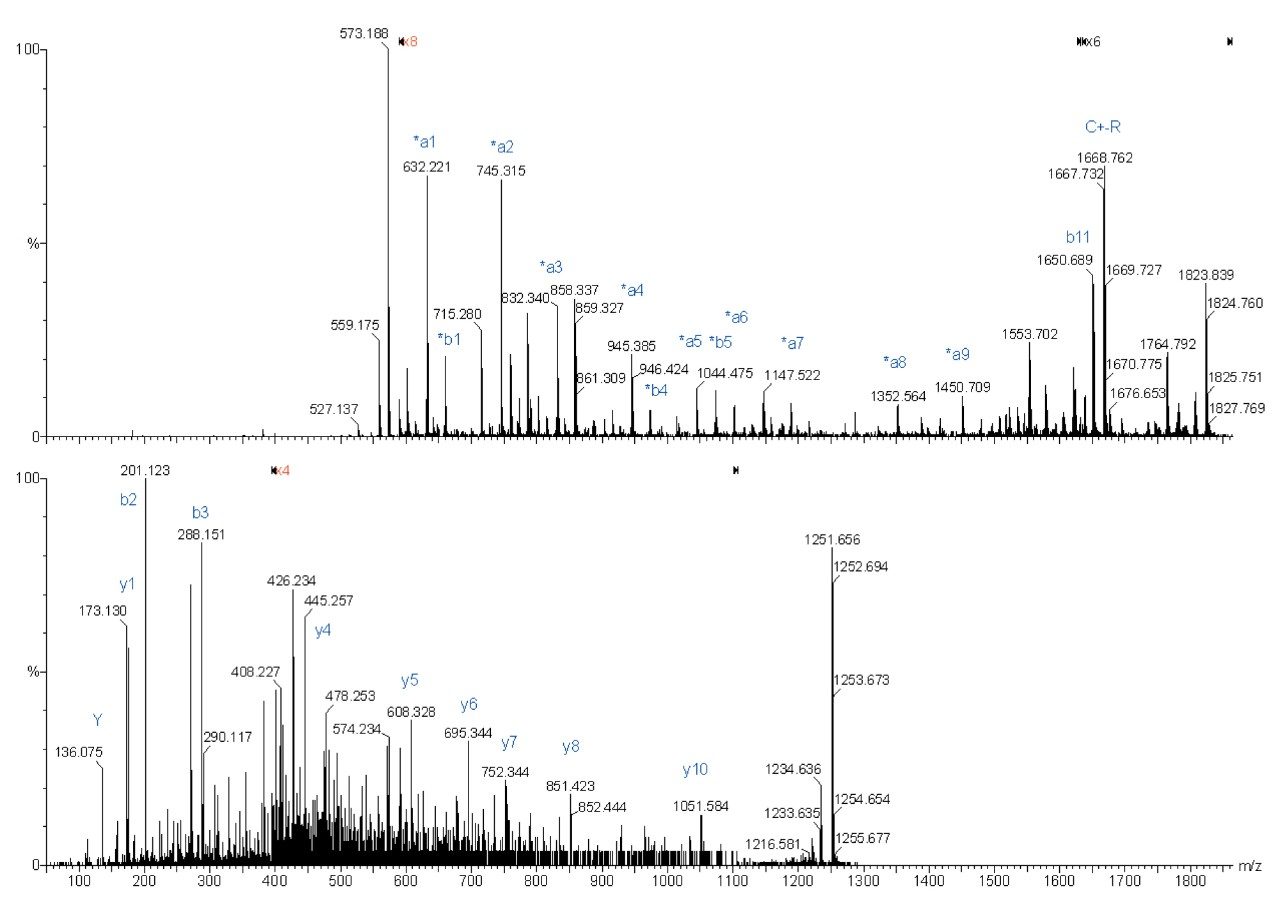
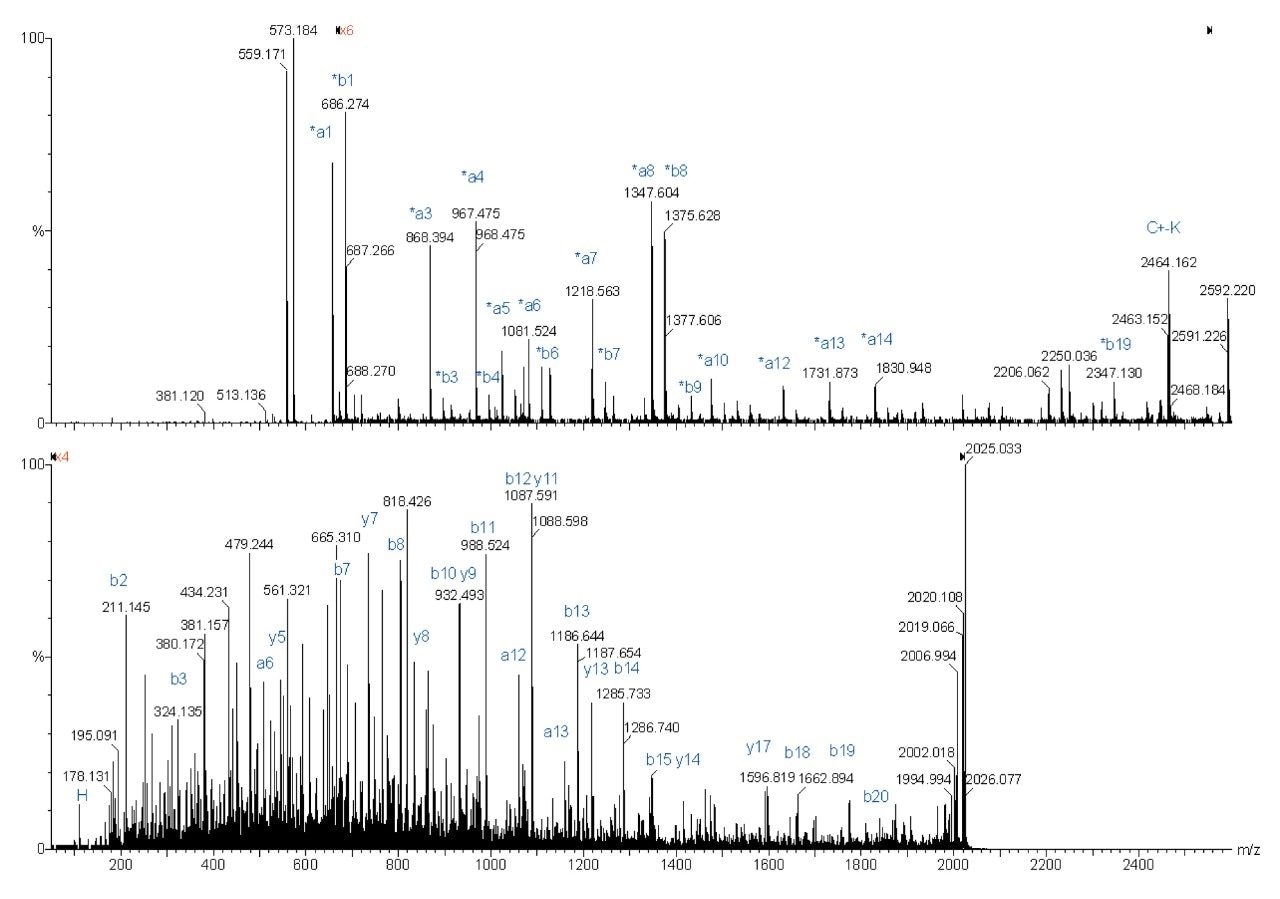
Sequence from y4 to y9 was found for the underivatized peptide, whereas almost a complete sequence was obtained with the derivatized peptide fragmentation.
The MS/MS spectrum of the underivatized peptide is much more difficult to interpret because of the larger number of different fragment ions. The derivatized peptide fragmentation gave almost exclusively *an and *bn fragment peaks. Also, more continuous sequence information was obtained.
720000958, September 2004