This is an Application Brief and does not contain a detailed Experimental section.
For research use only. Not for use in diagnostic procedures.
To utilize OneLab and the Andrew+ pipetting robot for automated metabolomic sample preparation.
Andrew+ pipetting robot streamlines the MTBE extraction method for lipid and small molecule analysis
Metabolomic studies often involve the analysis of small molecules and lipids in complex biological matrices. Due to the broad range of physiochemical properties of the compounds of interest in these studies, multiple extraction and analytical methods are required to obtain the desired coverage of molecules.1 Large scale epidemiological studies, which involve hundreds to thousands of biological samples, exaggerate these challenges leaving scientists to perform numerous pipetting steps and requiring large volumes of sample if multiple extractions are needed.2
Performing a single extraction methodology to obtain the broadest coverage of compounds is vital in increasing laboratory sample analysis throughput. A single liquid-liquid extraction protocol, however, is more complex and time consuming than conventional “dilute and shoot” extraction methods, which are favored in metabolomic studies.
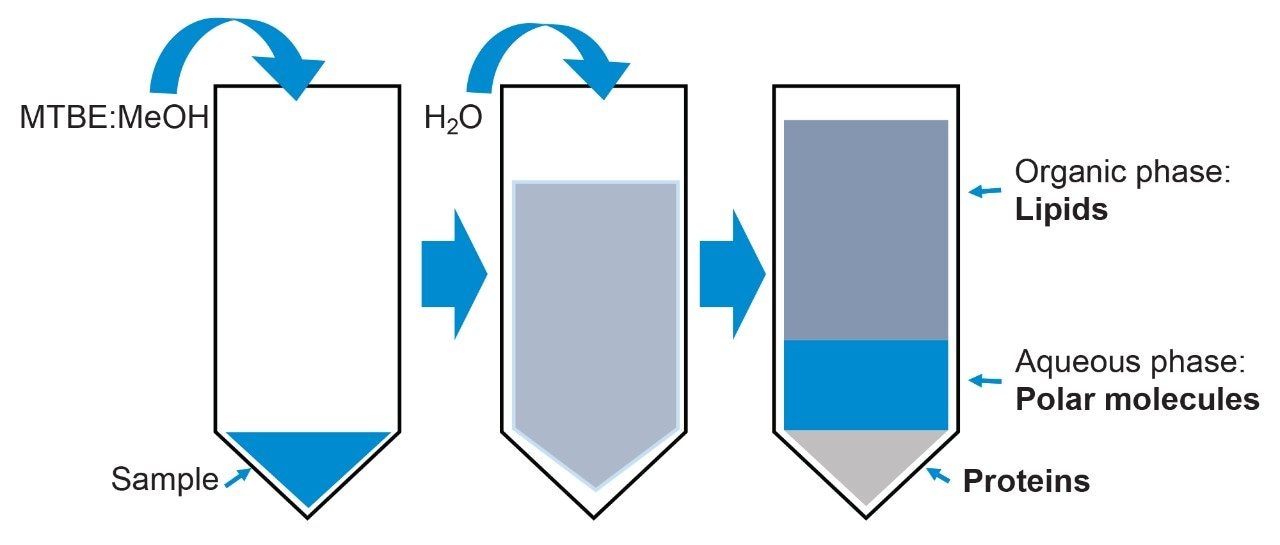
A single extraction protocol by Matyash et al3 outlines a liquid-liquid extraction method that is routinely employed in metabolomic laboratories for the extraction of lipids from plasma and tissue. Using methyl-tert-butyl ether (MTBE), methanol, and water, lipids are extracted into a layer of organic solvent following phase separation, while a lower aqueous phase in turn can be analysed for small polar molecules (Figure 1).
Automated liquid handling devices have been routinely employed in clinical laboratories to run validated extraction protocols on large batches of patient samples since the 90s, drastically reducing the need for manual human intervention.4,5 Still, many of these automated liquid handling platforms have large footprints, taking up considerable laboratory space greater than the space a scientist would use for the sample preparation. Additionally, programing the sample preparation protocols can be difficult with extensive training on the software being required and standalone PC’s required for each platform.
In this application brief, we describe how the Andrew+ pipetting robot and the OneLab protocol design and execution software has been used to perform the MTBE extraction method on human sputum samples, for metabolic phenotyping.
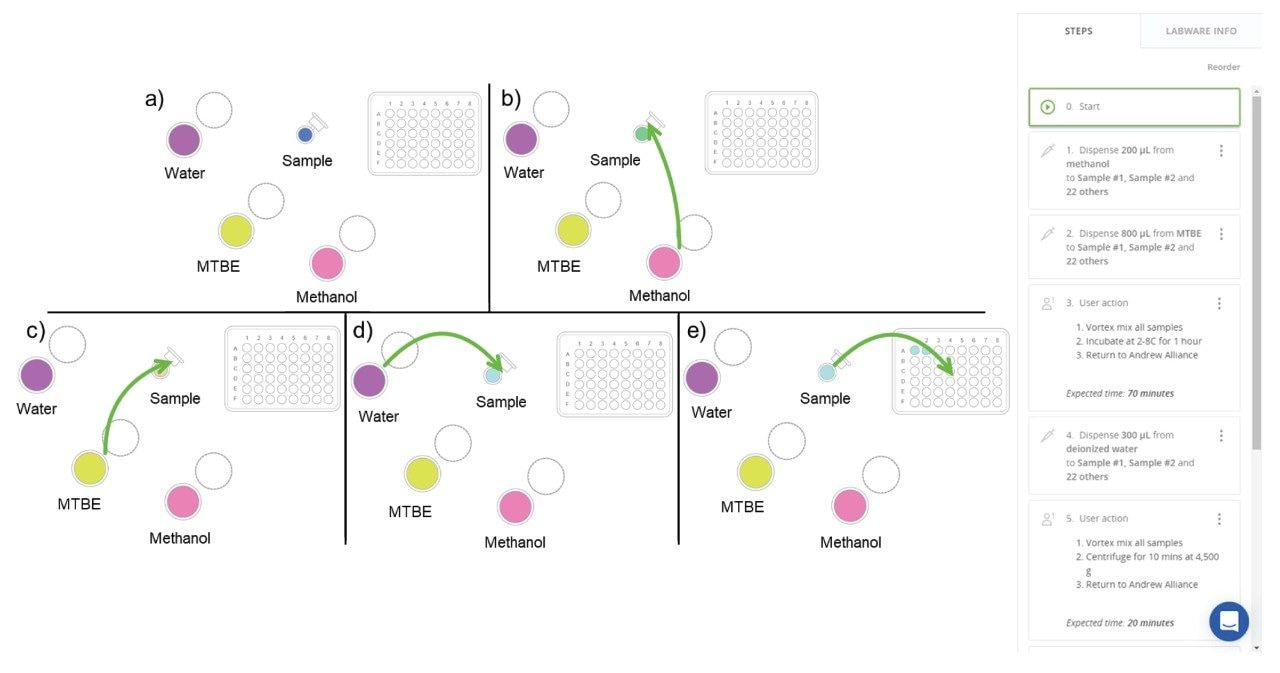
The use of automated pipetting robots helps regain much of the time lost to the sample preparation. Here, the Andrew+ pipetting robot was used to perform the MTBE liquid-liquid extraction method on human sputum samples. Each step of the extraction procedure was set up in the protocol design software, OneLab, where the appropriate labware for the sample volume was selected and added to the design page (Figure 2a). Figure 2b shows the first step of the extraction procedure where the Andrew+ robot pipettes 200 µL of methanol into each Eppendorf tube containing 100 µL of study sample. The second step in the protocol (Figure 2c) following the methanol addition, is the addition of 800 µL of MTBE to each sample. A manual user action step was incorporated into the protocol allowing dedicated time for each sample to be briefly vortex mixed prior to incubation at 2–8 °C for 1 hour, before returning them to the Andrew+ robot. Once returned, 300 µL of deionized water is pipetted into the sample Eppendorf tube (Figure 2d), and a further user action step was added for sample vortex mixing and centrifugation (4,500 x g for 10 minutes) to pellet out precipitated proteins and enable phase separation. The final steps of the OneLab protocol involved aliquoting the phase separated extraction into separate vials for the two phases (Figure 2e).
Following protocol design (or loading a OneLab protocol from the online library), the extraction method was selected to execute. Figure 3 shows the Andrew+ robot set-up required for the MTBE extraction protocol, displaying the domino position and the content of each domino ensuring the correct number of pipette tips, volume of extraction solvent, and study sample. After confirming the domino set up, the automated extraction procedure commenced with the extraction of 24 human sputum samples (100 µL) previously aliquoted into 1.5 mL Eppendorf tubes. Following completion of the protocol, the upper organic phase of the extract was dried down under nitrogen prior to reconstitution with isopropanol for LC-MS analysis. The lower aqueous phase was diluted with acetonitrile to achieve a composition of 1:1 aqueous sample to acetonitrile for HILIC analysis.

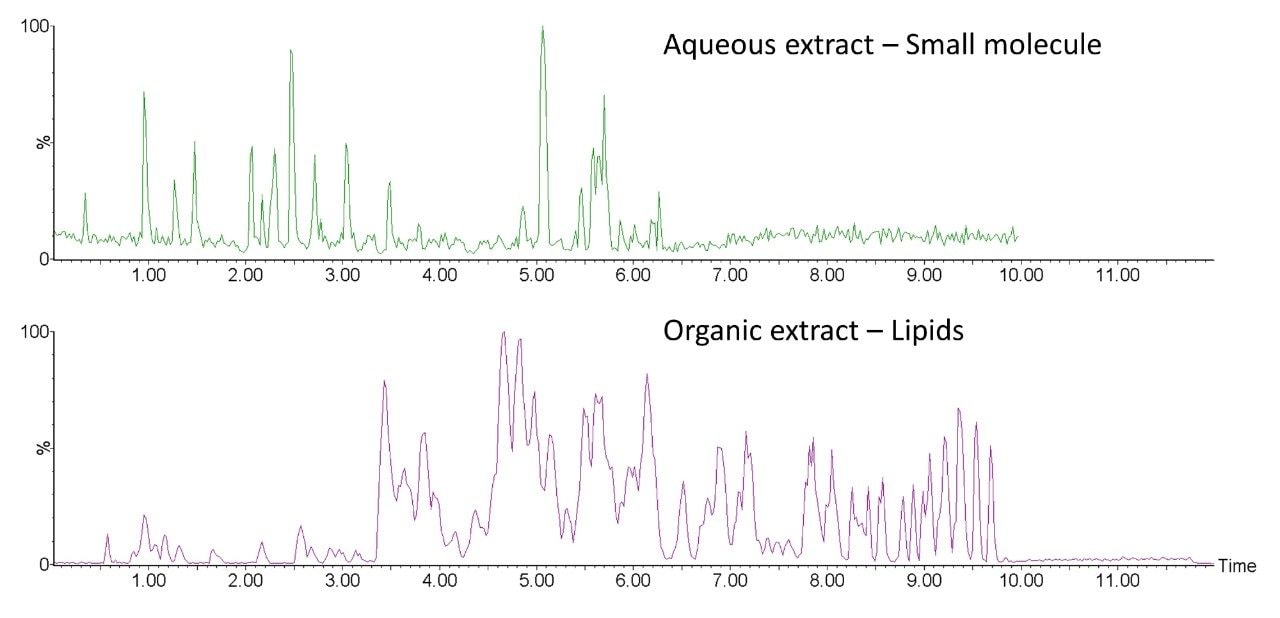
The resulting phases were individually analysed on a SYNAPT XS using a lipid reversed-phase chromatographic method (organic phase) and a HILIC based gradient method for the polar metabolites (aqueous phase). Figure 4 shows representative chromatograms of the extracted human sputum for both acquisition methods in positive ion mode, demonstrating a good recovery of polar metabolites (small molecule and polar lipids) alongside the non-polar lipids.
The MTBE lipid extraction method enables recovery of a broad range of lipids from biological matrices in addition to the extraction of polar metabolites in a lower aqueous phase, all from a single extraction procedure. The OneLab software allowed flexible and straightforward protocol design, with a variety of labware and sample volumes readily accommodated.
The Andrew+ robot demonstrated improved throughput with a reduction of ~30 minutes for the preparation of 24 biological samples when compared to a fully manual preparation procedure. The automated preparation consisted of fully automated pipetting (i.e. >1hr), thus allowing analysts to focus on other activities such as instrument set up.
720007303, July 2021