In this application note, we present a case study for the optimization of tryptic peptide mapping for a model protein such as phosphorylase B from Oryctolagus cuniculus (rabbit). Results on the identification of Lyc-C fragments of delta hemoglobin in the presence of other hemoglobin chains from human blood are shown as well. All data was acquired using a Waters LCT Premier oa-TOF MS in conjunction with ACQUITY UPLC.
Peptide mapping by means of LC/UV and/or LC-MS is an invaluable tool in analysis and quality control of biotherapeutic proteins. Peptide mapping has been widely used for purity testing, comparability testing and batch release of protein therapeutics. Through the implementation and now routine use of electrospray ionization and mass spectrometry (MS) over recent years, the field of protein chemistry has been revolutionized.
High resolution MS provided by bench-top orthogonal acceleration time-of-flight (oa-TOF) MS instrumentation has become widely used for protein chemistry analysis. MS yields important information about peptide sequence and can be used to detect and confirm post-translational modifications. The use of MS provides an additional dimension to that afforded by just chromatographic separation alone, and hence mass spectrometry is used in conjunction with conventional LC/UV peptide mapping to provide an extra level of analytical specificity. However, through the use of time-of-flight instrumentation, data quality can be further enhanced to give high accuracy information provided by exact mass measurement. The coupling of oa-TOF MS technology with the new advances in UltraPerformance LC (UPLC) now provides peptide chemists with a powerful and unique analytical tool giving unsurpassed analytical results.
In this application note, we present a case study for the optimization of tryptic peptide mapping for a model protein such as phosphorylase B from Oryctolagus cuniculus (rabbit). Results on the identification of Lys-C fragments of delta hemoglobin in the presence of other hemoglobin chains from human blood are shown as well. All data was acquired using a Waters LCT Premier oa-TOF MS in conjunction with ACQUITY UPLC.

The digest samples were analyzed by LC-MS. The samples – when required – were diluted with an aqueous 0.1% formic acid solution to an appropriate concentration. A Waters ACQUITY UPLC binary solvent manager was used to perform the UPLC separation with the column outlet directly connected to the source of the mass spectrometer. The analytical column was a 10 cm x 1 mm ACQUITY BEH C18 1.7 μm Column that was operated at a flow rate of 150 μL/min. Mobile phase A comprised of 0.1% formic acid in water and mobile phase B of 0.1% formic acid in acetonitrile. The gradient was from 5 to 30% in 15 min.
MS data was acquired using an LCT Premier oa-TOF mass spectrometer operating with positive electrospray ionization. The TOF analyzer was calibrated with a sodium formate solution. A 100 fmol/μL [Glu1]- Fibrinopeptide B solution was used as a ‘lockmass’ for exact mass measurement and was introduced into the mass spectrometer via the reference sprayer of the LockSpray ion source at a flow rate of 2 μL/min. LockSpray provides the ability to introduce a reference mass over long periods of time without interference from the analyte data. Having this capability provides a simple experimental setup for exact mass measurement whilst maintaining high quality data.
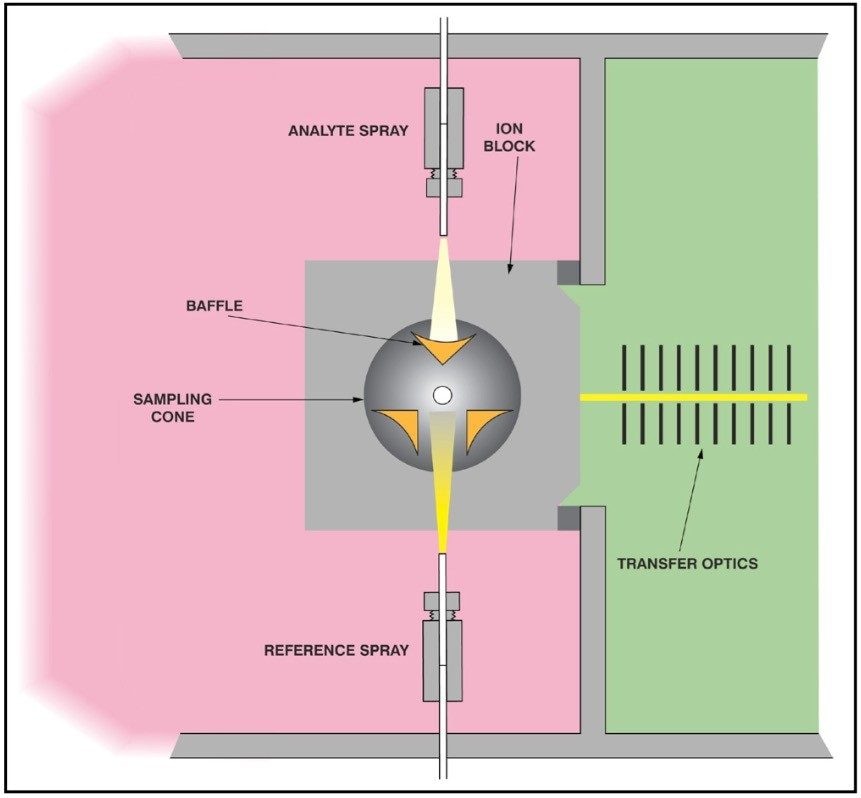
Peptide mapping mass accuracy feasibility study
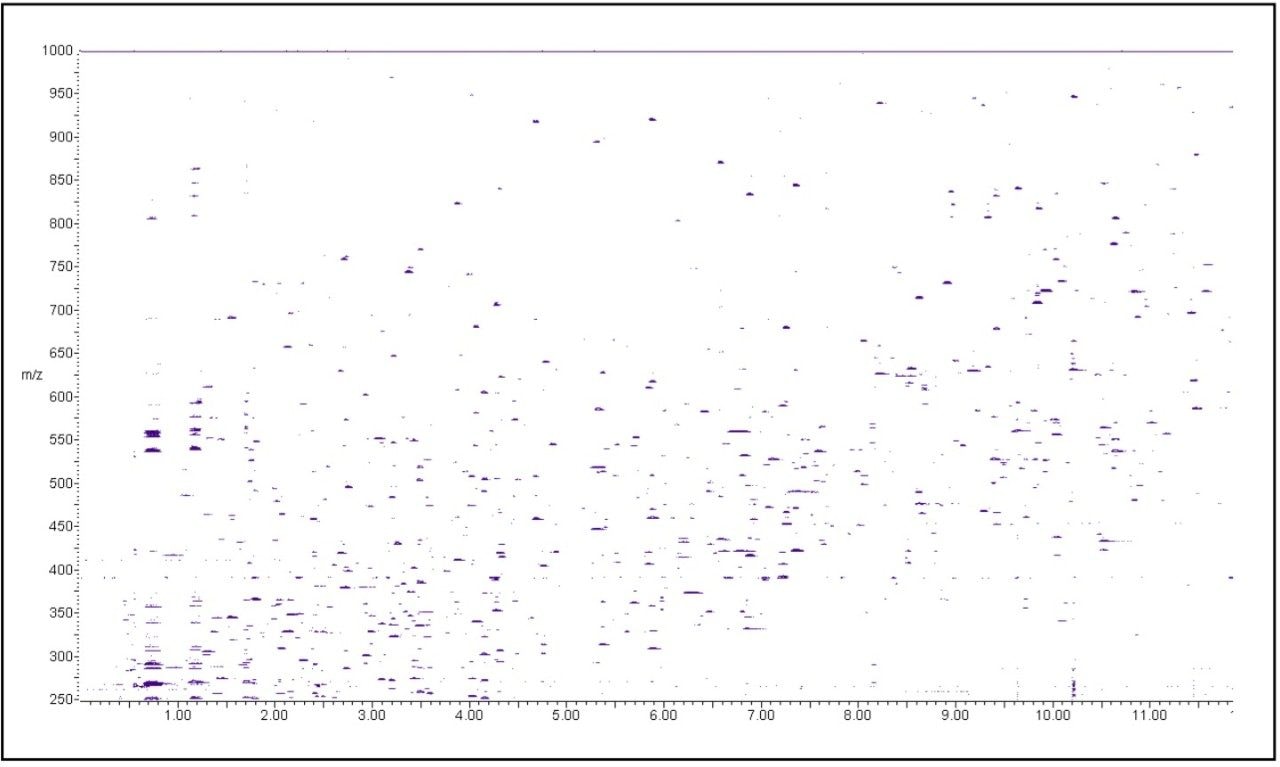
A 5 picomole amount of a MassPREP tryptic digest solution of phosphorylase B was injected on-column and used to optimize chromatographic resolution. The tryptic LC-MS map is shown in Figure 1, and shows a high degree of peak separation even with a relatively short run time. From the chromatogram, 35 unmodified peptides were selected and mass measured to determine the RMS mass measurement error. The latter was conducted automatically with the all file accurate mass measure option of MassLynx 4.1 Software. Missed cleavages were not considered for the mass accuracy calculations and the results are summarized in Table 1.
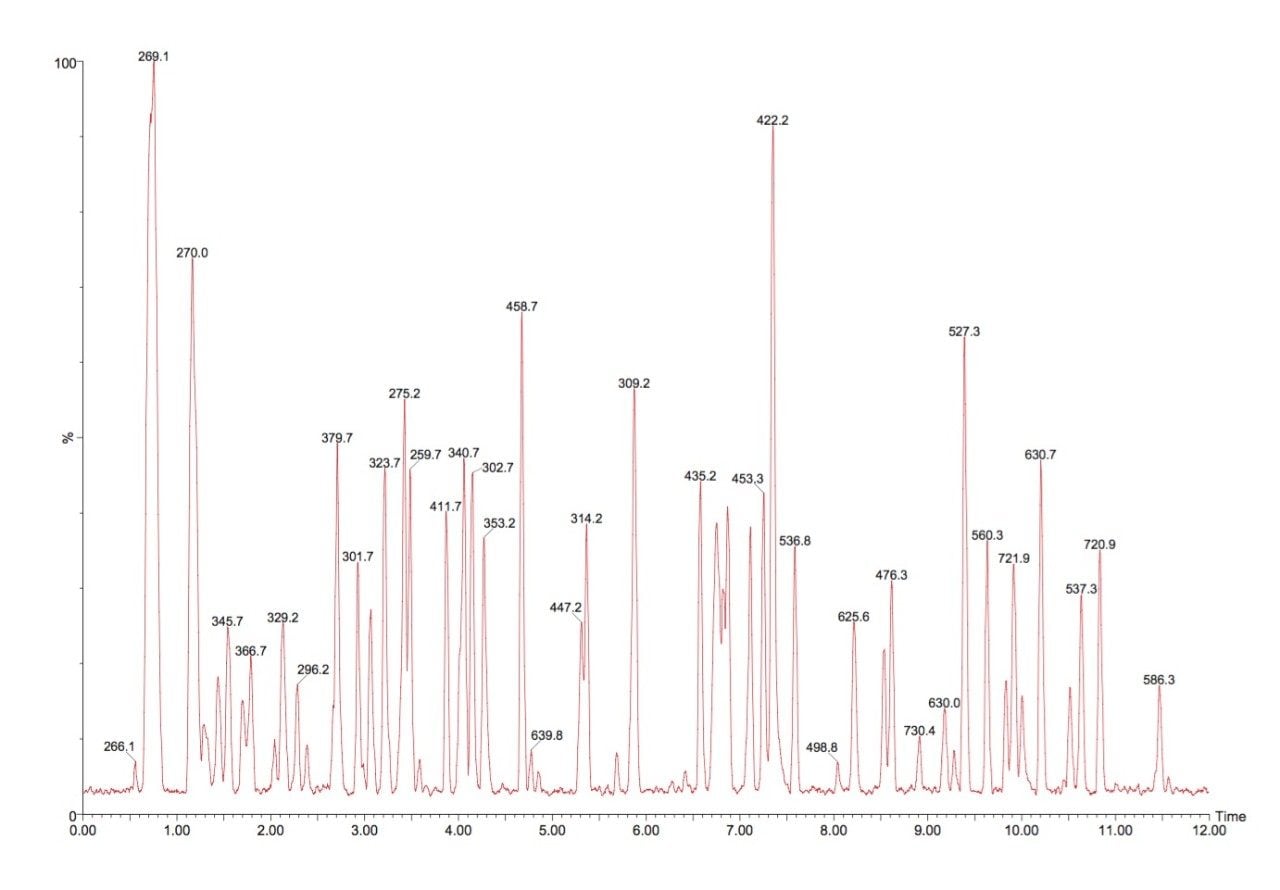
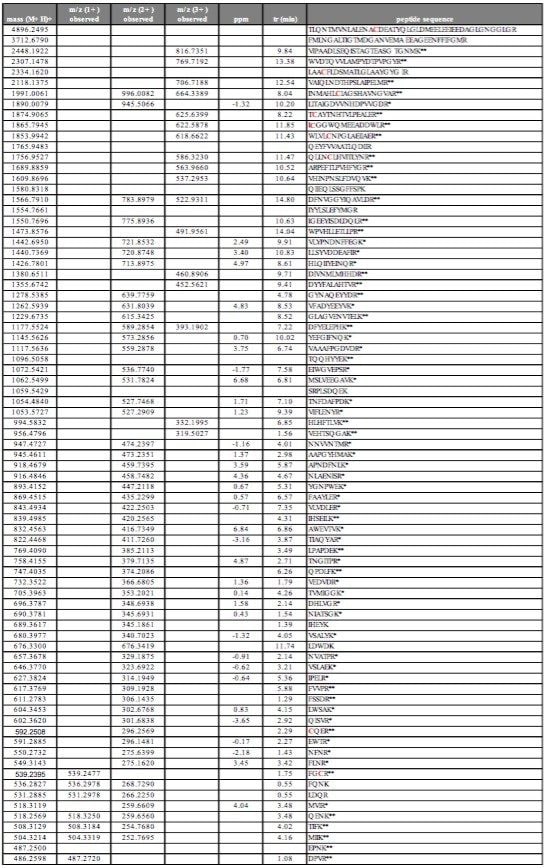
Shown in Table 1 are the theoretical masses of the singly charged tryptic precursor ion, the observed mass, the obtained mass error in ppm, retention time and amino acid sequence. The RMS mass error for all the 35 selected peptides was found to be 2.9 ppm. With the use of UPLC, peak widths are narrower than with conventional HPLC due to the use of a 1.7 μm particle size. The average chromatographic peak width during this analysis was approximately 4 s at 10% height and 2 s at half height but even with these peak widths, good peak definition and exact mass measurement was possible due to the fast data acquisition capabilities of the LCT Premier.
Additional peptides were identified by extracting all theoretical masses of the digest of phosphorylase B, which yielded a total of 70 identified peptides. This equals a total amino acid sequence coverage of 75% and a theoretical coverage based on m/z range of 90%. The contour plot – ion intensity vs. m/z and retention time - is given in Figure 2 to illustrate the ion density of the analyses.
As an alternative data processing procedure, all spectra across the peptide map were combined and single point mass measured at 50% of the gradient time, i.e. ~ 6 min. The combined and mass corrected spectrum was deconvoluted with MaxEnt3 to yield a monoisotopic, singly charged and de-isotoped spectrum, which was subsequently submitted for a peptide mass fingerprint search against the SwissProt database. In this instance, a 10ppm mass tolerance, carbamidomethylation of cysteine residues and one missed cleavage site were considered. 5 different phosphorylase isoforms were identified of which the rabbit protein homolog exhibited the greatest identification probability. 47 peptides were identified with an RMS mass error of better than 5.0 ppm and displayed amino acid sequence coverage of 49.0%.
Comprehensive well-characterized biopharmaceutical applications require complete base line separation of all peptides. To maximize resolution and sensitivity in peptide mapping with UPLC, it is recommended to use flow rates on the order of 100–200 μL/min with 2.1 mm columns and 50 μL/min with 1 mm columns.1 The presented UPLC-oa-Tof System offers the flexibility to either obtain peptide maps at enhanced speed or enhanced sensitivity and chromatographic resolution.
Lyc C delta chain peptide mapping
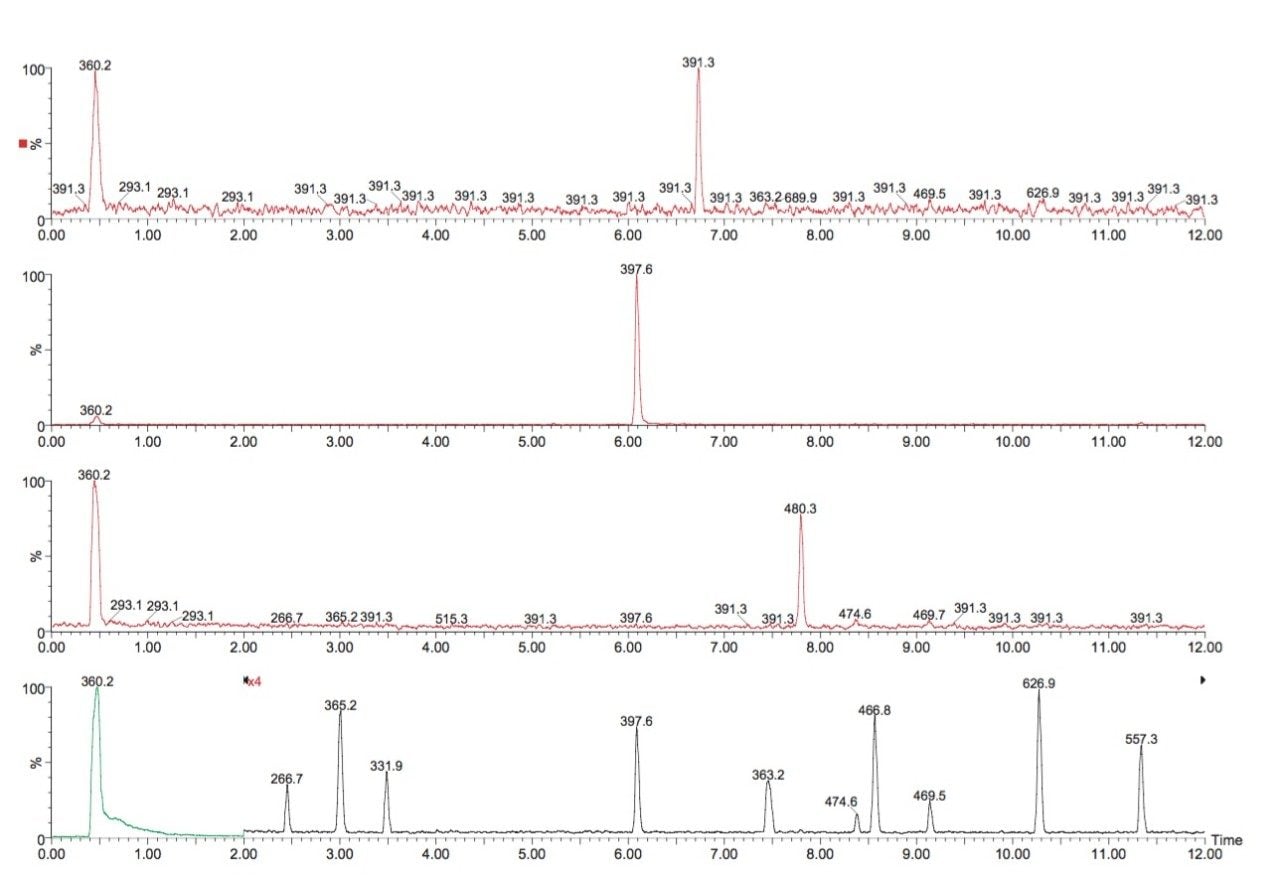
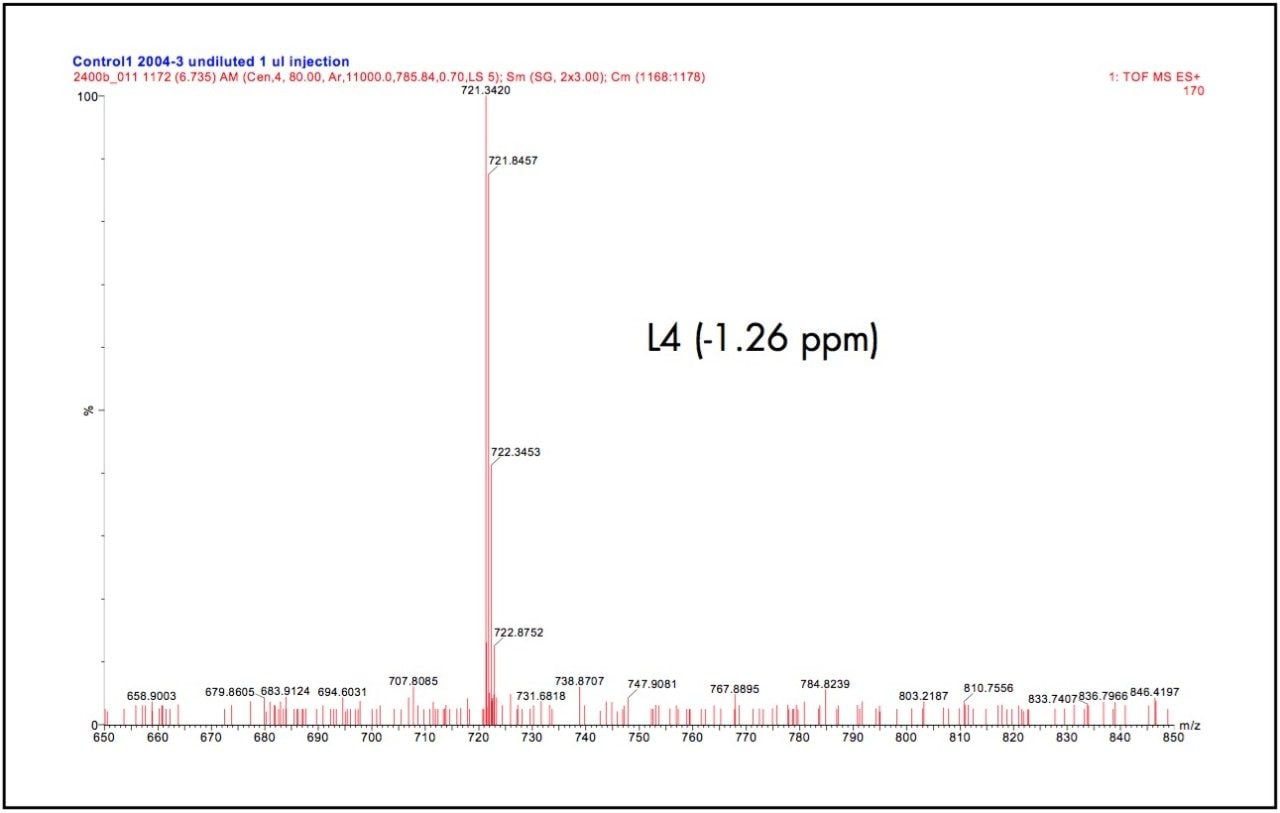
1 μL of digested human serum sample was injected on column, which corresponded to approximately a 30pmol load of total protein digest. The main hemoglobin components in human serum comprise of the alpha and beta chain and to a lesser extent the delta and gamma chains. The LC-MS chromatograms of the Lys-C digest of this sample is shown in Figure 3(a). The peptides of interest originate from the delta chain and are present at low concentrations resulting in reduced electrospray signal intensities. Due to the low ion currents produced, statistics challenged the exact mass measurement analysis. Nevertheless, the RMS error for the three peptides of interest was found to be better than 4.6 ppm, even at very low ion currents. Shown in Figure 3(b) is one of the obtained peptide spectra (L4).
The aim of this application note was to show that the use of UPLC coupled directly to time-of-flight mass spectrometry has a significant benefit for the analysis of peptides. The facility of exact mass measurement provided by time-of-flight MS greatly enhances the analytical answer produced and provides an extra level of confidence when determining analytes of interests. In addition to sensitive full spectral acquisitions, exact mass measured data is produced routinely providing a high degree of analytical specificity. Hence, the method benefits from the orthogonal separation on m/z of co-eluting species by means of oa-Tof mass analysis. Through the use of LockSpray, the practical aspects associated with exact mass measurement are made easy with excellent reliability. With the advent of UPLC, separations of peptide mixtures can be carried out faster whilst still maintaining excellent separation power and thus giving a high level of peptide coverage. The combination of both UPLC and TOF provides an unsurpassed level of analytical power that greatly enhances peptide mapping studies.
ECMP, Laboratoire de Spectrometrie de masse bioorganique, Strasbourg, France is kindly acknowledged for providing the Lys-C digest of human hemoglobins.
720001830, August 2006