For research use only. Not for use in diagnostic procedures.
In this application note, novel RP-UPLC separations are performed using a Waters ACQUITY UPLC System with Charged Surface Hybrid (CSH) chemistry. The combination of sub-2 μm particle size with an optimized liquid chromatography system and novel chemistry allows for a significantly improved RP method that maximizes the performance of these particles and is optimized for the analysis of complex lipid mixtures.
Lipids play key roles in human health. Alterations in lipid levels have been associated with the occurrence of various diseases, including cardiovascular diseases, diabetes, cancer, and neurodegenerative diseases.1 Advances in LC-MS have allowed lipids to be studied with greater sensitivity and specificity, alleviating the effects of co-eluting compounds and isobaric interference, and allowing low abundance lipids to be more readily detected.1
Conventional mass spectrometric analysis of lipids is often performed by direct infusion, or reversed-phase (RP) / normal-phase (NP) HPLC.2-5 However, each of these methods faces its own challenges.
With direct infusion, chromatographic separation of lipids is not performed prior to injection into the mass spectrometer. This method of sample introduction gives rise to ion suppression and it does not allow for separation of isobaric lipids, which can complicate the resultant analysis, necessitating deconvolution, and compromising the sensitivity of the method. In order to fully explore the lipidome, a technique of sample introduction into the mass spectrometer that minimizes these issues is needed.
NP chromatography allows separation of lipids by class but often suffers from long elution times, is difficult to handle due to the volatility and toxicity of the mobile phase, and proves challenging for ionization and introduction into mass spectrometry.6 Recent work in HILIC chromatography overcomes many of these issues.7
Traditional RP methods similarly suffer from extensive elution times and the quality of the resulting chromatography is relatively poor. Peak capacity and resolution are compromised in a typical analysis and it is not unusual to see peaks widths > 30 seconds,5 which ultimately results in poor sensitivity and difficulty in characterization.
In this application note, novel RP-UPLC separations are performed using a Waters ACQUITY UPLC System with Charged Surface Hybrid (CSH) C18 chemistry. The combination of sub-2-µm particle size with an optimized liquid chromatography system and novel chemistry allows for a significantly improved RP method that maximizes the performance of these particles and is optimized for the analysis of complex lipid mixtures.
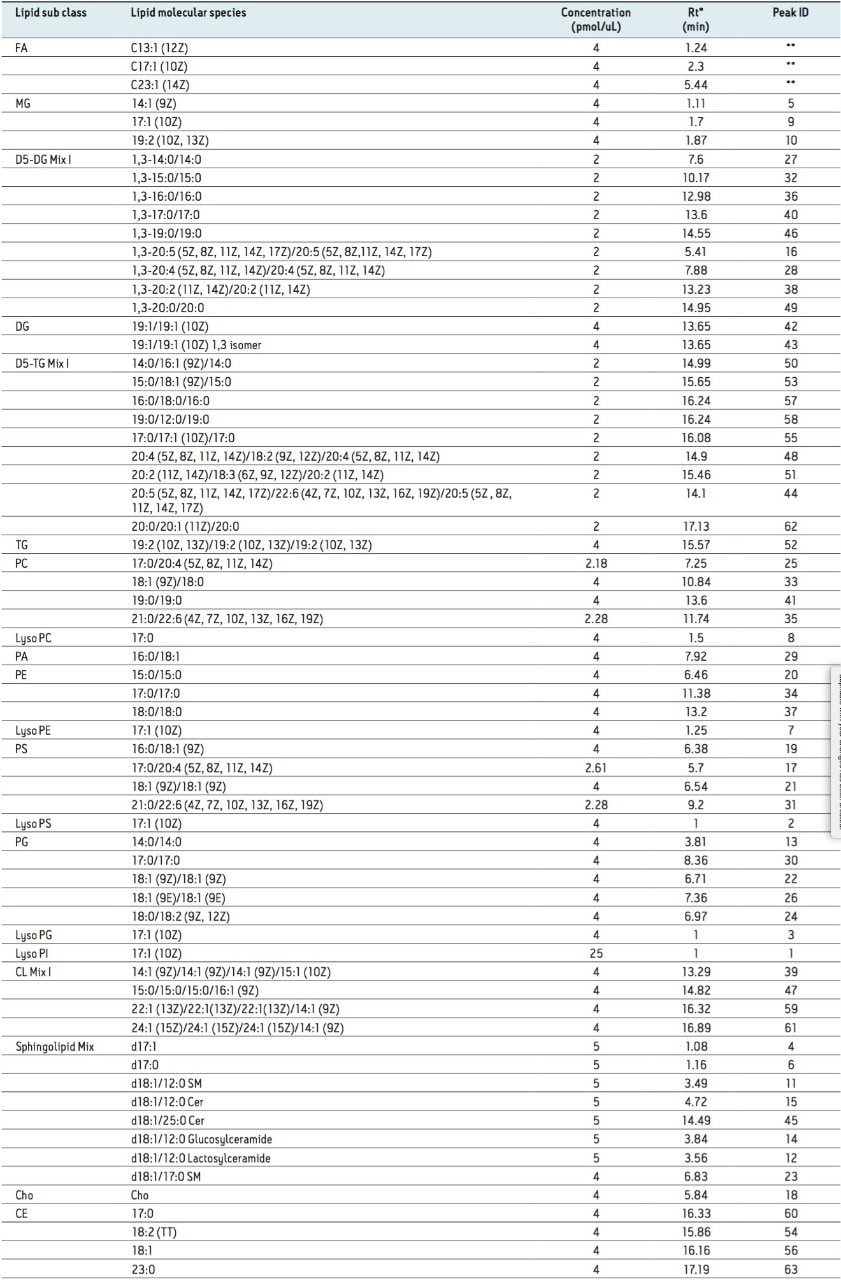
Lipid standards were purchased from Avanti Polar Lipids and Nu-Chek Prep. The standards were diluted prior to analysis in isopropanol/acetonitrile/water (2:1:1, 250 µL). The list of lipid standards analyzed and other detailed information is provided in Table 1.
A total lipid extract from bovine liver was purchased from Avanti Polar Lipids. The extract was prepared by making a 5 mg/mL stock solution in chloroform/methanol (2:1). A working 0.1 mg/mL solution was then prepared by diluting the stock solution with isopropanol/acetonitrile/water (2:1:1).
Rat plasma (25 μL) from Equitech-Bio, Inc. was extracted with 100 µL of chloroform/methanol (2:1); this solution was then allowed to stand for 5 min at room temperature, followed by vortexing for 30 s. After centrifuging (12,000 g, for 5 min at 4 °C) the lower organic phase was collected in a new vial and evaporated to dryness under vacuum. Immediately prior to analysis the lipid extract was diluted with isopropanol/acetonitrile/water (2:1:1, 250 μL ).
|
LC system: |
ACQUITY UPLC |
|
Column: |
ACQUITY UPLC CSH C18 2.1 x 100 mm, 1.7 μm |
|
Column temp.: |
55 °C |
|
Flow rate: |
400 μL/min |
|
Mobile phase A: |
Acetonitrile/water (60:40) with 10 mM ammonium formate and 0.1% formic acid |
|
Mobile phase B: |
Isopropanol/acetonitrile (90:10) with 10 mM ammonium formate and 0.1% formic acid |
|
Injection volume: |
5 μL |
|
Time (min) |
% A |
% B |
Curve |
|---|---|---|---|
|
Initial |
60 |
40 |
Initial |
|
2 |
57 |
43 |
6 |
|
2.1 |
50 |
50 |
1 |
|
12 |
46 |
54 |
6 |
|
12.1 |
30 |
70 |
1 |
|
18 |
1 |
99 |
6 |
|
18.1 |
60 |
40 |
6 |
|
20 |
60 |
40 |
1 |
|
Mass spectrometer: |
SYNAPT G2 HDMS |
|
Acquisition mode: |
MSE |
|
Ionization mode: |
ESI positive/negative |
|
Capillary voltage: |
2.0 KV (for positive) |
|
|
1.0 KV (for negative) |
|
Cone voltage: |
30 V |
|
Desolvation temp.: |
550 °C |
|
Desolvation gas: |
900 L/Hr |
|
Source temp.: |
120 °C |
|
Acquisition range: |
100 to 2000 m/z |
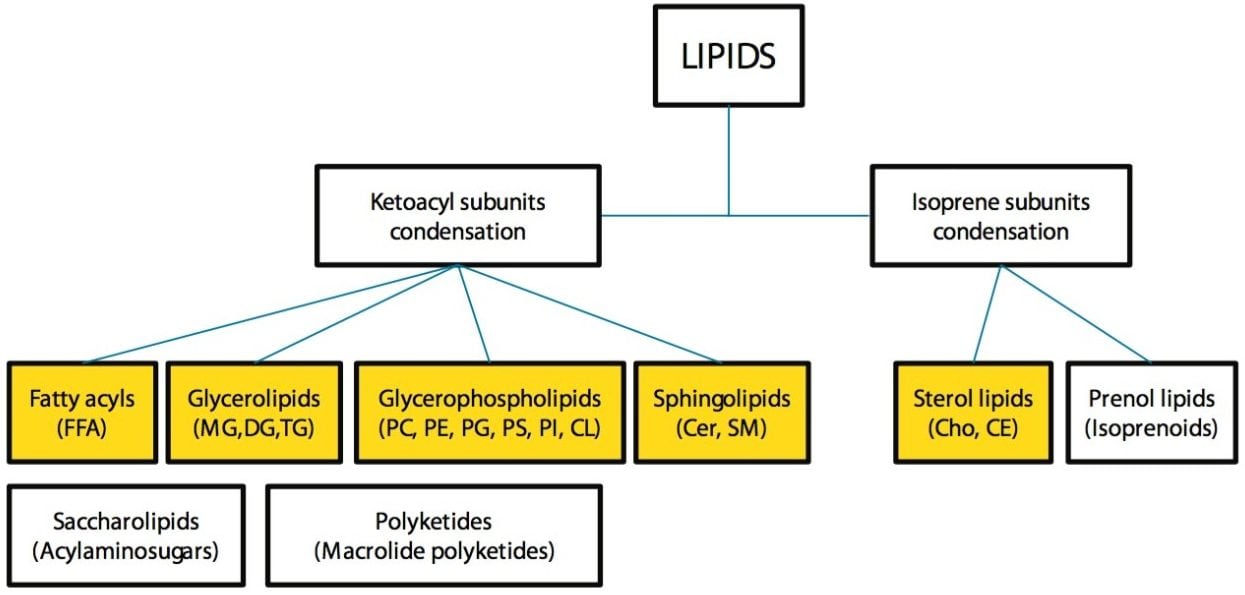
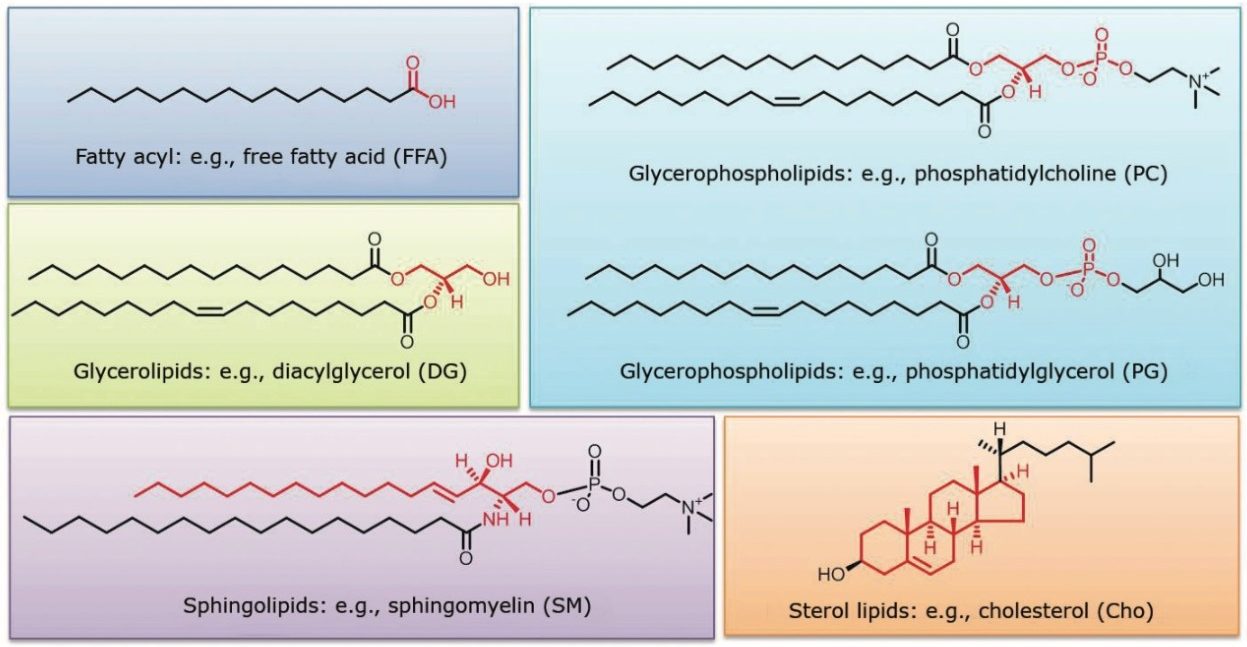
The UPLC RP method employed for this analysis showed improved separation of both inter and intra class lipids over traditional HPLC RP methods. Figures 1 and 2 show the lipid classes based on Lipid MAPS classification and representative structures for major lipid categories respectively. Highlighted in yellow are the classes that are included in the standard mix, which reflect the most abundant lipids present in animal tissues.
The resolution, sensitivity, and speed of analysis were significantly increased compared with HPLC. The CSH chemistry’s charged surface is believed to interact with the lipids on column in a unique manner. Due to the diverse chemical nature of lipids, from highly polar to highly non-polar, a different mechanism of retention from traditional RP columns was observed.
The additional speed allows for large sample sets to be analyzed efficiently, while the added resolution and sensitivity increase peak purity. This provides added confidence in the assignment of a lipid or class of lipids while allowing identification to be made at lower concentrations.
Our initial analysis focused on the separation of a mixture of 67 lipid standards, as shown in Table 1.
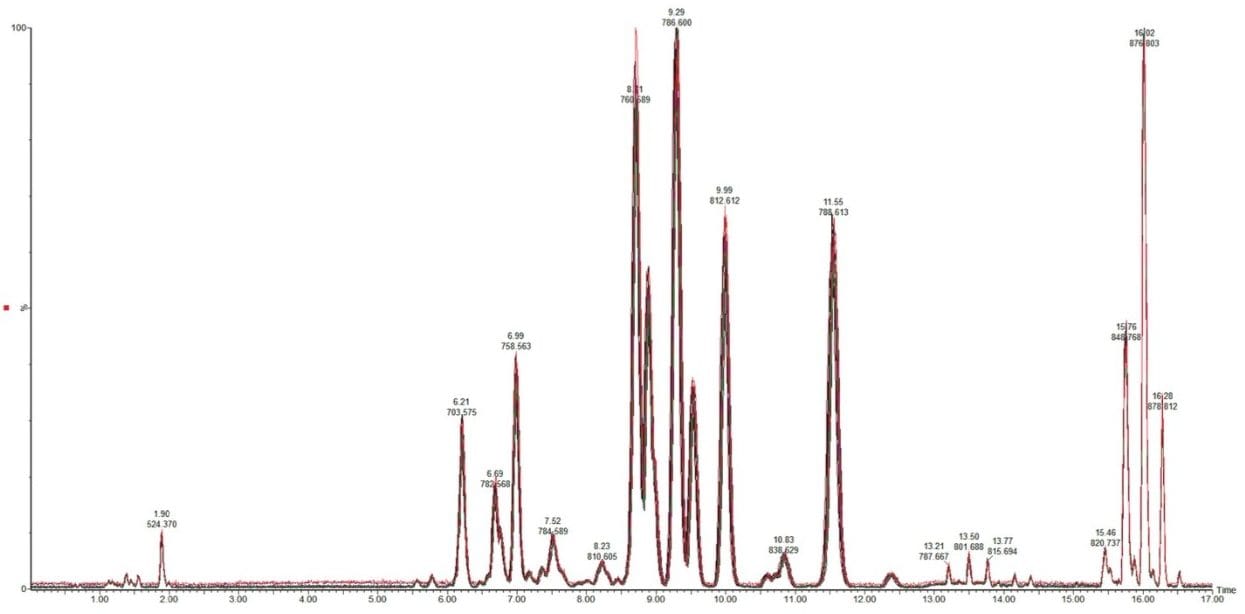
The resulting base peak intensity chromatogram can be seen in Figure 3.
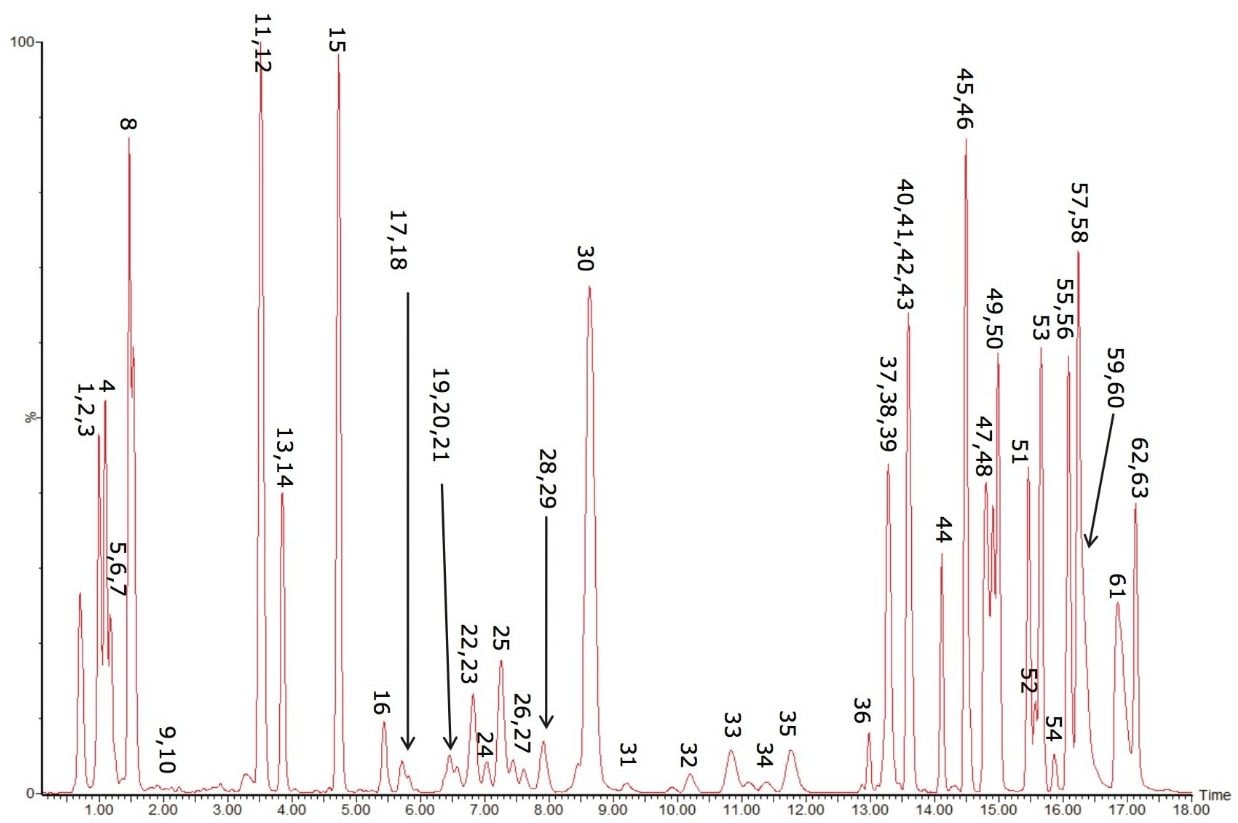
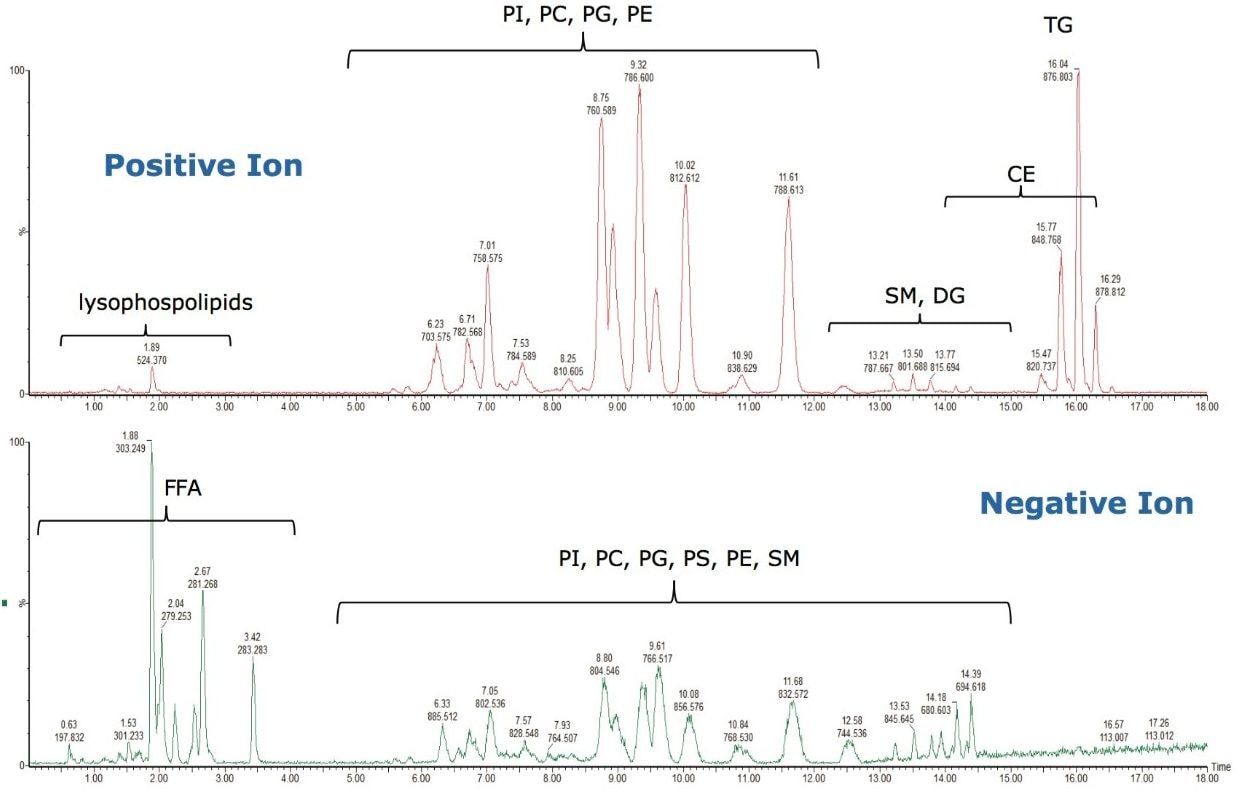
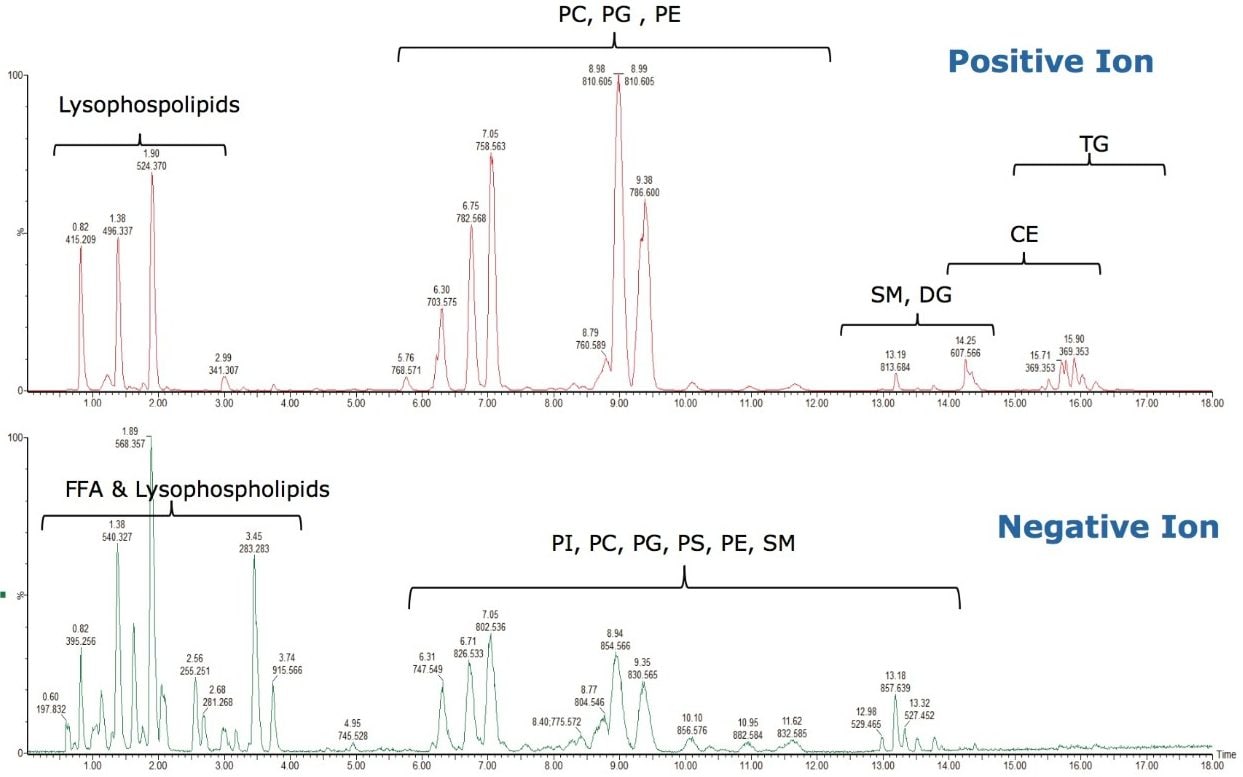
As can be seen in Figure 3, several interesting observations were made during the course of developing this method. First, we have shown previously that the phophatidylcholines (PC) typically co-elute with sphingomyelins (SM) during a RP separation.8 Using CSH technology C18, an enhanced separation of these classes was observed. This is especially useful when coupling with mass spectral detection as both classes fragment to give a phosphocholine ion at m/z 184.074, the common head group. Also, in complex biological matricies, SM are less abundant than PC species, leading to ion suppression effects that affect the efficiency of detection of minor species. Figures 4 and 5 show the analysis of total lipid extracts from bovine liver and rat plasma respectively.
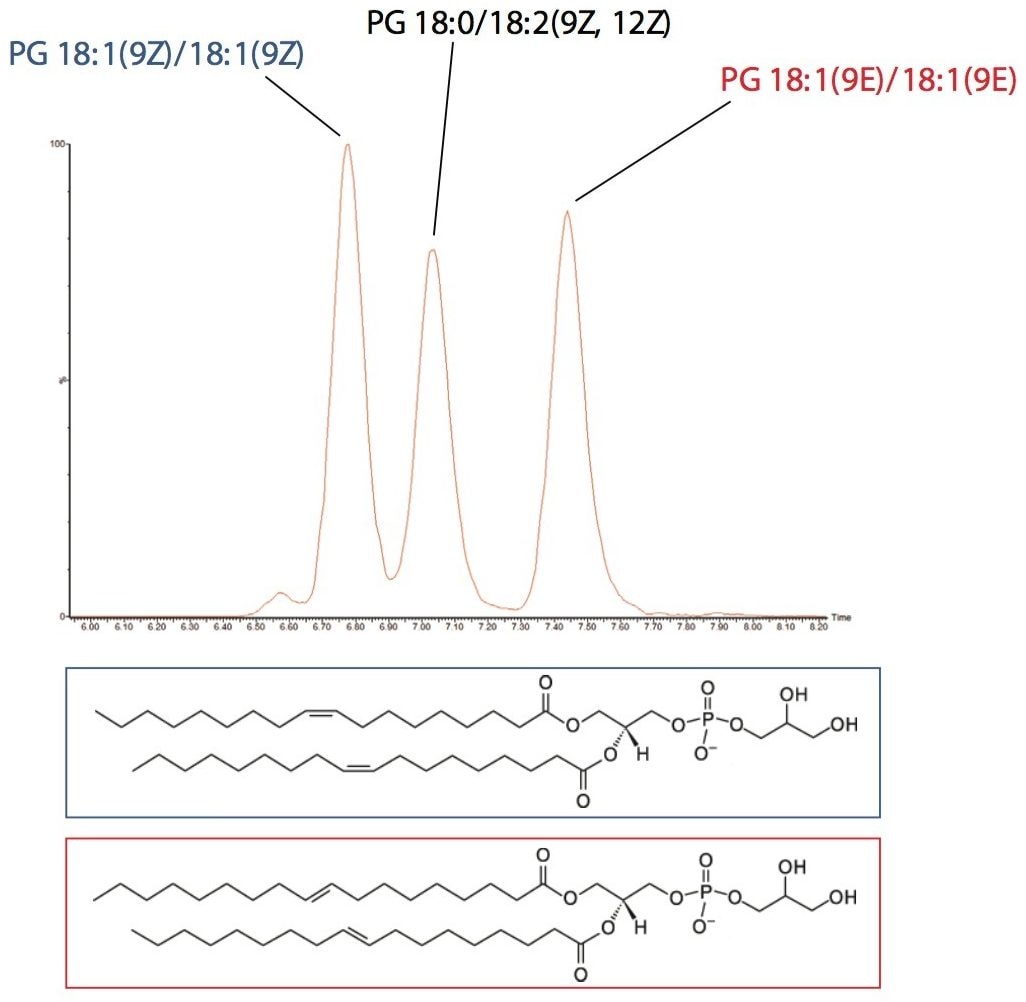
In this analysis the ability of UPLC and CSH Technology to differentiate between structural cis (Z) and trans (E) isomers was also observed. The separation of cis and trans isobaric phosphatidylglycerol (PG) species, such as PG 18:1(9Z)/18:1(9Z) and PG 18:1(9E)/18:1(9E) were easily separated. In addition, structural isomers such as PG 18:1(9Z)/18:1(9Z) versus PG 18:0/18:2(9Z, 12Z) were resolved, as shown in Figure 6. This information would typically not be available using an infusion or traditional HPLC methods.
To test the applicability of this novel chromatographic method in real biological samples, we analyzed total lipid extracts from bovine liver and rat plasma. Using the CSH C18 ACQUITY UPLC System, we were able to separate the major lipid classes with high resolution and sensitivity, which improved the detection of low abundant lipid species, as shown in Figures 4 and 5. Notably, the CSH C18 ACQUITY UPLC System presented excellent retention time reproducibility from multiple injections of a bovine liver extract (%RSD < 0.136; n=20), as shown in Figure 7. This is especially useful for lipidomic analysis, which requires the comparison of a large number of LC-MS chromatograms deriving from multiple sample sets.
The use of UPLC with CSH C18 column described in this method provides clear improvements over other NP and RP traditional HPLC methods. It also marks an improvement over the UPLC methods highlighted in previous application notes for lipid analysis.8 This method offers a robust and reliable approach for lipid analysis.
720004107, September 2011