This is an Application Brief and does not contain a detailed Experimental section.
Size-exclusion chromatography (SEC) is commonly used to support the development of biotherapeutic protein products to evaluate protein self-association and fragmentation. However, in-process purification and formulation development samples can often result in the target protein being in solutions that can form precipitates which can greatly decrease SEC column life. Here we demonstrate the utility of the BioResolve SEC mAb, 200 Å, 2.5 µm Guard Column to provide effective protection for an analytical BioResolve SEC, 2.5 µm, 7.8 x 300 mm Column, and having minimum impact on resolution. In addition, we demonstrate the predicted increased tolerance of 2.5 µm particle size columns towards samples with suspended protein precipitates versus a column with a 1.7 µm particle size.
Protein self-associated forms, or high molecular weight species (HMWS) are routinely assessed as a critical quality attribute (CQA) in biotherapeutic protein preparations as they can impact both the safety and efficacy of treatment.1 SEC remains one of the predominant methods for the assessment of protein self-association due to the reliability and sample throughput of the method.2 Although, it is critical that the veracity of the SEC measurement be confirmed by complementary methods such as analytical ultracentrifugation (AUC).3 There has also been an increase in the measurement of protein fragment levels, or low molecular weight species (LMWS) by SEC, particularly for monoclonal antibody based therapeutics.4 The increased demand for greater sample throughput has necessitated the use of more efficient, smaller particle size (sub-2-µm) ultra-performance SEC (UP-SEC) columns. UP-SEC columns are also packed in smaller 4.6 mm inner diameter (I.D.) columns that require LC systems with significantly lower dispersion versus larger particle sizes packed in 7.8 mm I.D. high-performance SEC columns (SE-HPLC).5
In addition to being less tolerant of LC system dispersion, UP-SEC columns are also more demanding of guard column performance in order to maintain optimal resolution. As a result, high efficiency guard columns are needed for UP-SEC. A previous publication showed the ability to obtain monoclonal antibody resolutions using a 2.5 µm particle size, 7.8 mm I.D. SE-HPLC column that were comparable to those obtained on an UP-SEC column with only a modest increase in analysis time.6 For the same reasons that a 7.8 mm I.D. SE-HPLC column can be effectively used on LC systems with greater dispersion, the demands on SE-HPLC guard column efficiency are lower. Additionally, due to the larger particle size of the HP-SEC column, we observed an increased tolerance for samples with suspended protein precipitates versus a column with a 1.7 µm particle size as would be predicted. These attributes should certainly be considered when the protein samples to be tested may have uncertain levels of particulates.
The effectiveness of the BioResolve SEC mA, 200 Å, 2.5 µm Guard (p/n: 186009443) to protect a 7.8 x 300 mm BioResolve SEC mAb Column (p/n: 186009441) from the injection of suspended protein precipitates was evaluated. In addition, a comparison of the susceptibility to the injection of precipitated protein was made between the BioResolve SEC mAb, 2.5 µm Guard and the ACQUITY Protein BEH UPLC SEC, 200 Å, 1.7 µm Guard (p/n: 186005793). Precipitated protein was generated by stressing the BEH200 SEC Protein Standard Mix (p/n: 186006518, reconstituted in 500 µL of water) at 80 ºC on an orbital shaker set at 800 RPM for 30 minutes. The performance of the separation was evaluated using an expired sample of Erbitux (cetuximab) as provided in a liquid formulation at 2 mg/mL.
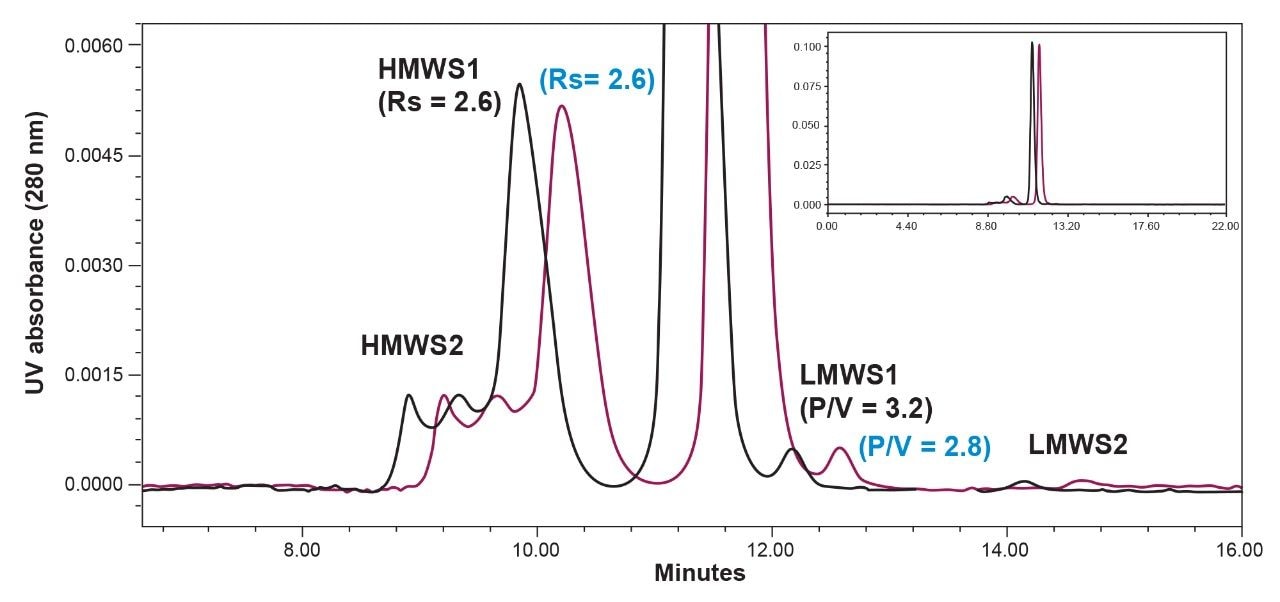
The impact that the BioResolve SEC mAb Guard has on the separation of cetuximab was evaluated (Figure 1). The experimental details are provided in the caption of Figure 1. We observe a predicted increase in retention time and minimal change in the quality of the separation. The resolutions (USP) for HMWS1 were equivalent and a slight decrease in the peak-to-valley ratio (P/V) of LMWS1 with the guard in place was observed. Given that LMWS1 is present at a 0.5% level, the modest decrease in P/V is likely the result of the increase in the extent of low level tailing due to dispersion effects.7
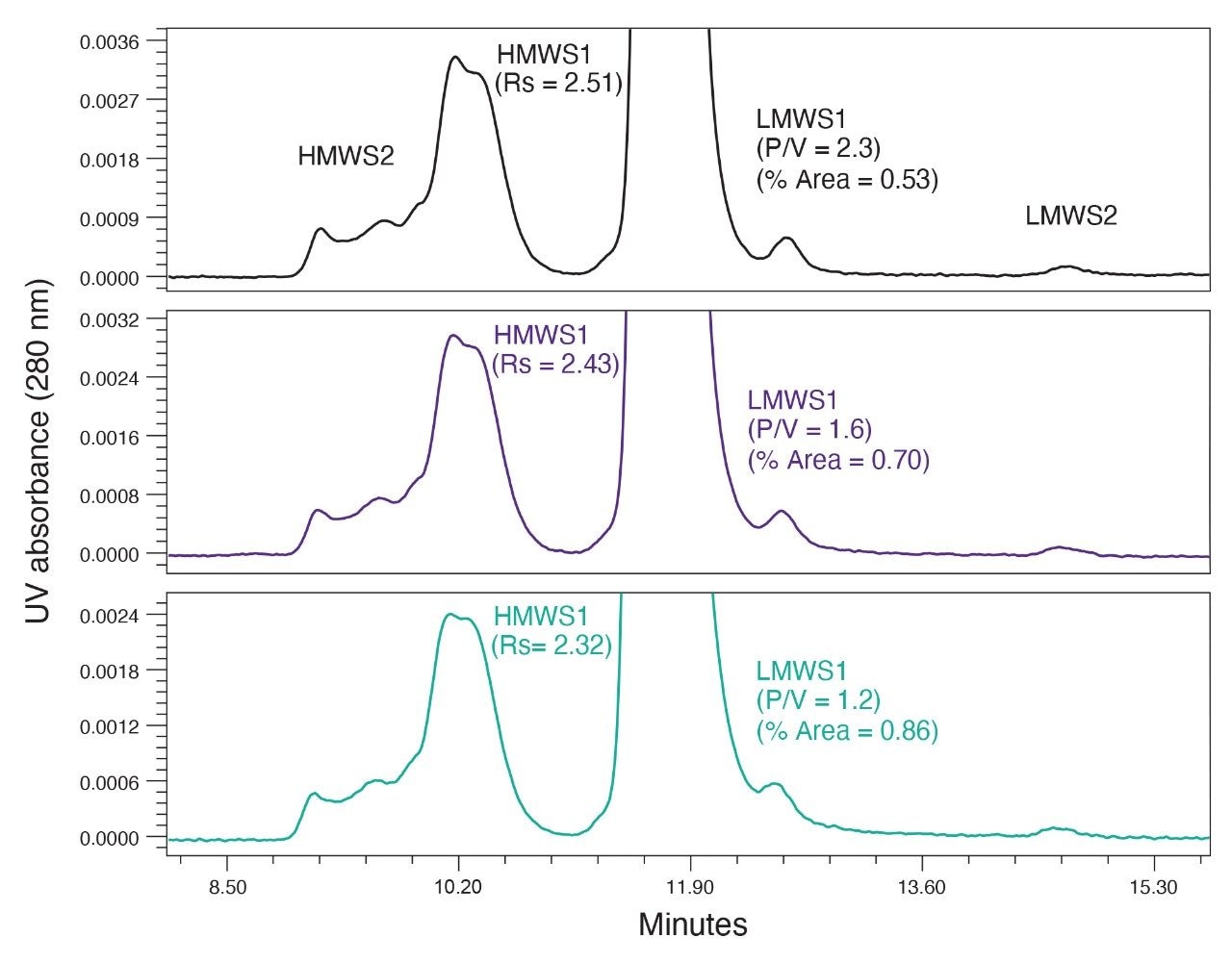
Next, a series of 14 µL injections of the stressed protein standard were made onto the column with the guard in place, and the separation of cetuximab was evaluated after every 20 injections (Figure 2). Over the course of 40 stressed protein standard injections we observed a steady decrease in the resolution of HMWS1. Additionally, we observed some changes in the chromatographic profile of the HMWS1 and HMWS2. It was noted that the percent peak areas of the HMWS2 and HMWS1 changed throughout these studies which is predominately the result of the high levels and instability of these self-associated forms. The high-levels of HMWS2 and HMWS1 are the result of freeze-thaw cycles, which are not recommended when this product is to be used clinically. As a result, these values are not reported.
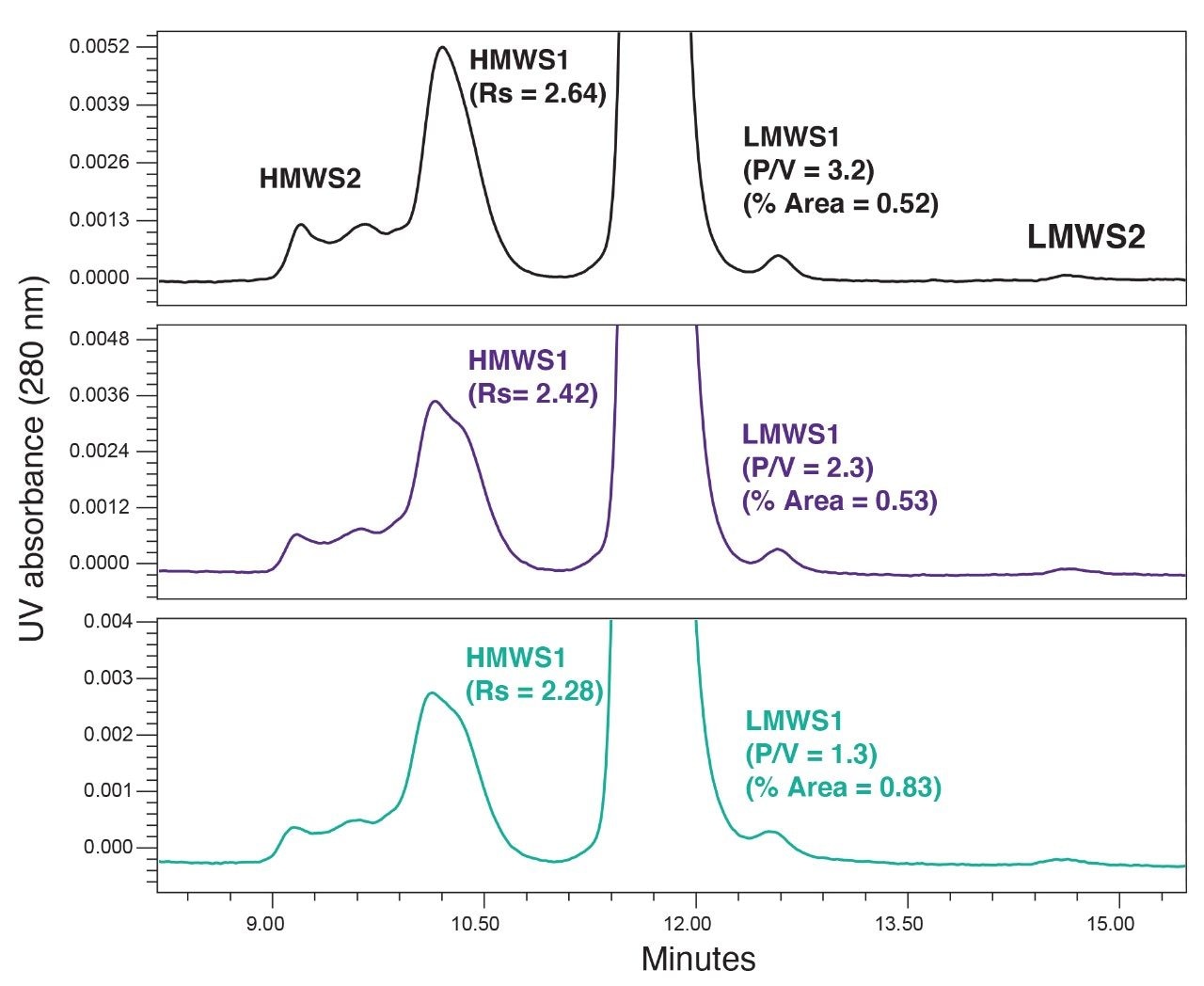
A decrease in the P/V of LMWS1 is also observed and more significantly, a 60% increase in the percent peak area of LMWS1 was measured. This is likely the result of increased low-level tailing of the monomer peak. This is important to note as the level of LMWS1 may be considered as a CQA for mAb-based therapeutics.
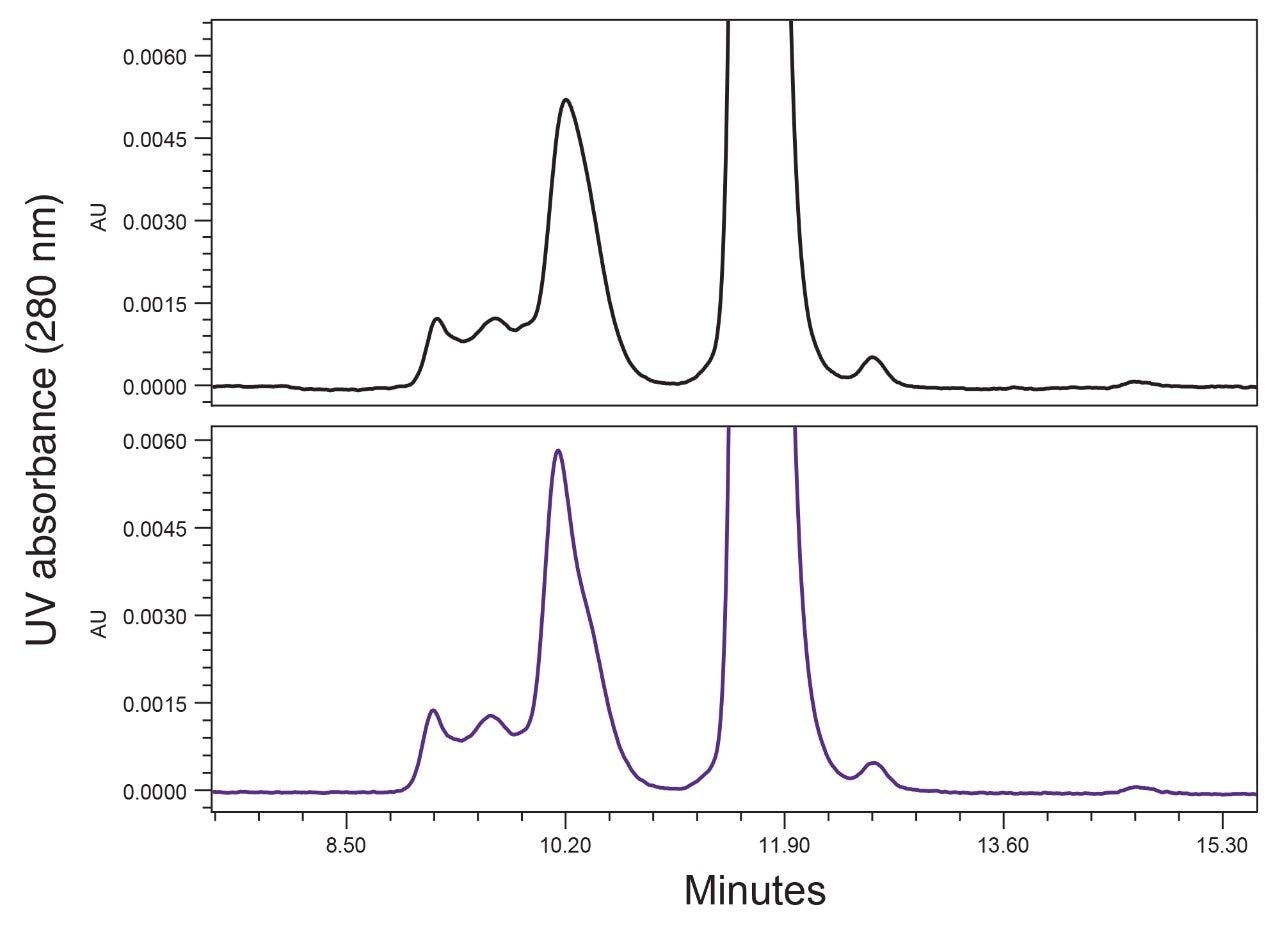
The fouled guard was then replaced, and the separation of cetuximab was reevaluated (Figure 3). As shown, the chromatographic performance was improved substantially. While the resolution of HMWS1 and the P/V of LMWS1 were not fully restored to their initial levels, most of the previous performance was recovered, and the significant increase observed in the percent peak area of LMWS1 was reversed. These data indicate that the BioResolve SEC mAb Guard can effectively extend the life of the analytical column when analyzing samples containing precipitated proteins.
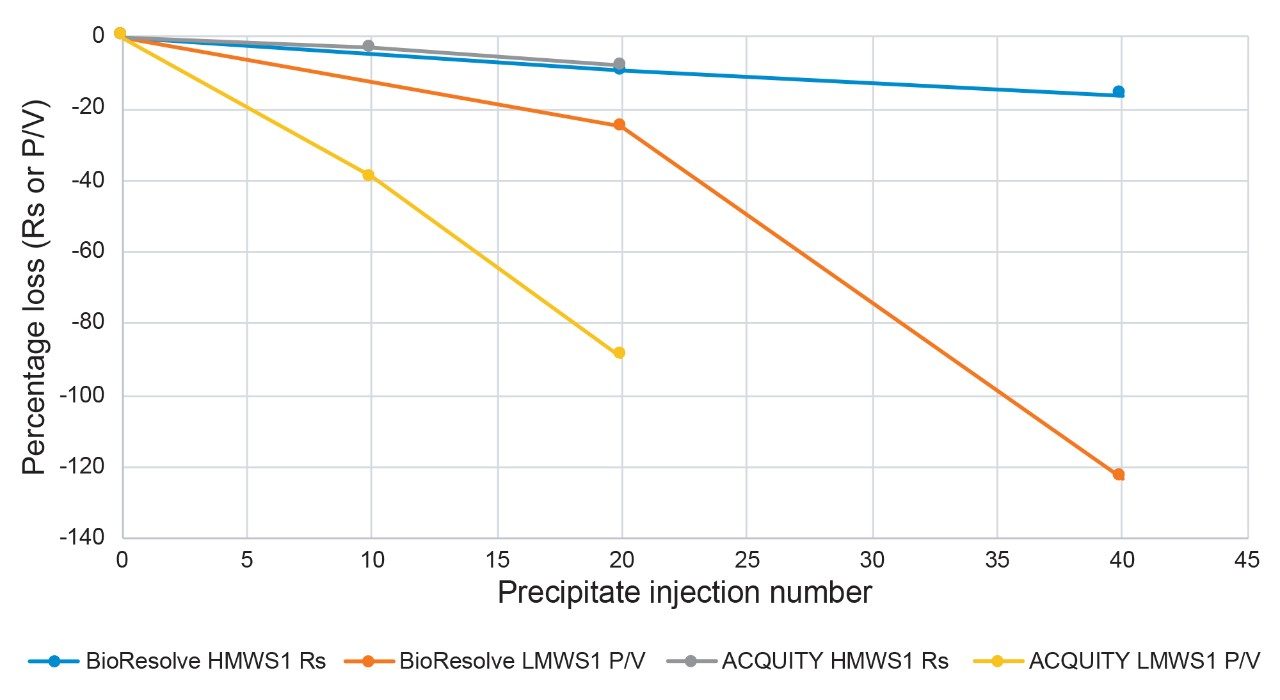
As a final study, the rate of fouling of the BioResolve SEC mAb, 2.5 µm, 4.6 x 30 mm Guard was compared to that of the equivalently sized ACQUITY UPLC Protein BEH SEC, 1.7 µm Guard while using the same 2.5 µm particle size, 7.8 x 300 mm analytical column. For this study, smaller injections (5 µL) of the stressed protein standard were made on the 1.7 µm particle size guard, since it is intended to be used with a 4.6 mm I.D. analytical column, and the separation of cetuximab was evaluated after every 10 injections (Figure 4). We observed the rate of loss of initial performance was comparable for the resolution of HMWS1 and that of the P/V of LMWS1 appeared to be greater over the initial 20 injections of the precipitated protein sample. Taking into account that the two guards were of the same I.D. and length, and the injection volumes were almost three times larger for the 2.5 µm particle size guard, these results clearly indicate that a 2.5 µm particle size SEC column is significantly less prone to fouling by precipitated proteins versus an SEC column packed with 1.7 µm particles. This is predicted since a larger particle size would result in reduced filtration of these particulates. It would also be predicted that the impact that a fouled guard would have on these separations would be greater on a 4.6 mm I.D. versus a 7.8 mm I.D. analytical column due to the larger peak volumes generated by the latter.
These results demonstrate that the use of a BioResolve SEC mAB Guard (p/n: 186009443) to protect a BioResolve SEC mAb Column can be beneficial when analyzing both HMWS and LMWS in process development samples that may contain varying levels of precipitated protein.
It is also shown that 2.5 µm particle size HP-SEC columns are significantly more resistant to performance loss when injected with precipitated proteins versus an UP-SEC column packed with 1.7 µm particles.
As a final note, while the use of a guard column can be beneficial, sample preparation to remove particulates via filtration or more preferably centrifugation to minimize potential speciation should also be considered whenever possible.
We would like to acknowledge the contributions of Lavelay Kizekai and Steve Shiner for providing the mobile phase and columns used in this study, and Bill Warren and Pam Iraneta for their review of the manuscript.
720006955, August 2020