HRMS Quantitation of Oligonucleotides Using Xevo G2-XS QTof Mass Spectrometer
Abstract
This application demonstrates the quantitative and qualitative capabilities of the Xevo G2-XS QTof and its suitability for oligonucleotide bioanalysis. We also demonstrate comparable performance with the Xevo TQ-XS Triple Quadrupole Mass Spectrometer for routine use.
Benefits
- High resolution Xevo G2-XS QTof Mass Spectrometer is suitable for routine quantitation of oligonucleotides such as GEM 91 (Trecorvirsen)
- ACQUITY Premier UPLC System and ACQUITY Premier Oligonucleotide C18 Column technologies improve oligonucleotide chromatographic recovery, LLOQs, and linear dynamic range
Introduction
Interest in utilizing HRMS for quantification has increased for a variety of factors including increased mass range, alternative selectivity (relative to tandem SRM/MRM), and most importantly, its ability to collect additional qualitative data. HRMS instruments are capable of quantitative performance that is comparable to that of tandem (triple) quadrupole instruments while providing modern bioanalytical labs additional selectivity options and analytical modes of acquisition (including DIA, DDA, and targeted modes). In the area of oligonucleotides, HRMS in particular has been increasingly utilized.1
In this application note, we outline the performance capabilities of the Xevo G2-XS QTof HRMS platform for oligonucleotides as well as compare typical bioanalytical figures of merit to the triple quadrupole platforms. Xevo TQ-XS Triple Quadrupole performance and methodology has been previously described in detail a previous application note.3
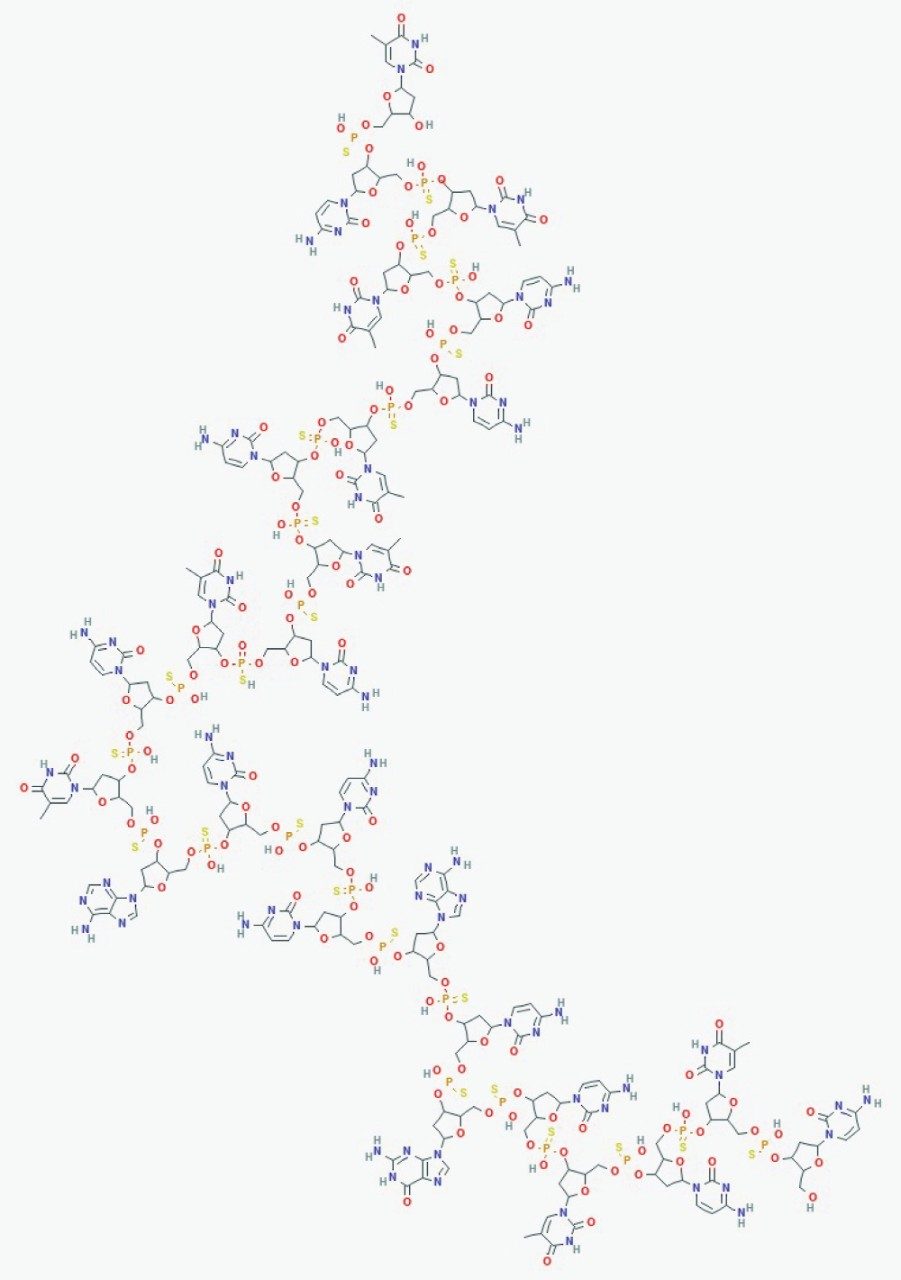
To demonstrate the capabilities of HRMS, detection and quantification of oligodeoxythymidines standards and GEM91, a fully phosphorothioated antisense oligonucleotide (Figure 1) were performed on the Xevo G2-XS QTof and compared to Tandem Quadrupole Mass Spectrometers using the ACQUITY Premier UPLC System.4
Experimental
Stock solutions of the Waters MassPREP Oligonucleotide Separation Technology (OST) Standard (p/n: 186004135) and the oligodeoxynucleotide phosphorothioate (GEM91; custom synthesized by Nitto Denko Avecia, Milford, MA) were prepared at 1 mg/mL and 0.1 mg/mL respectively in proteinase (RNAse) free water. LC-MS samples were prepared by diluting stock solutions in a reagent consisting of mobile phase A and SPE eluate solvent at 50:50 volume ratio. A 250 ng/mL concentration of GEM 132 was used as IS to evaluate/quantify GEM 91.
For additional methodology, please refer to the application note titled “Improved Oligonucleotide SPE-LC-MS Analysis Using MaxPeak High Performance Technology” (https://www.waters.com/content/dam/waters/en/app-notes/2020/720007019/720007019-en.pdf).
Gradient
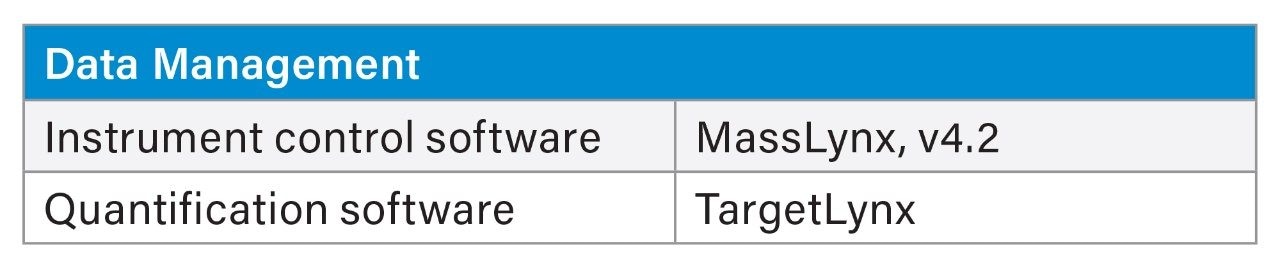
MS Conditions
Results and Discussion
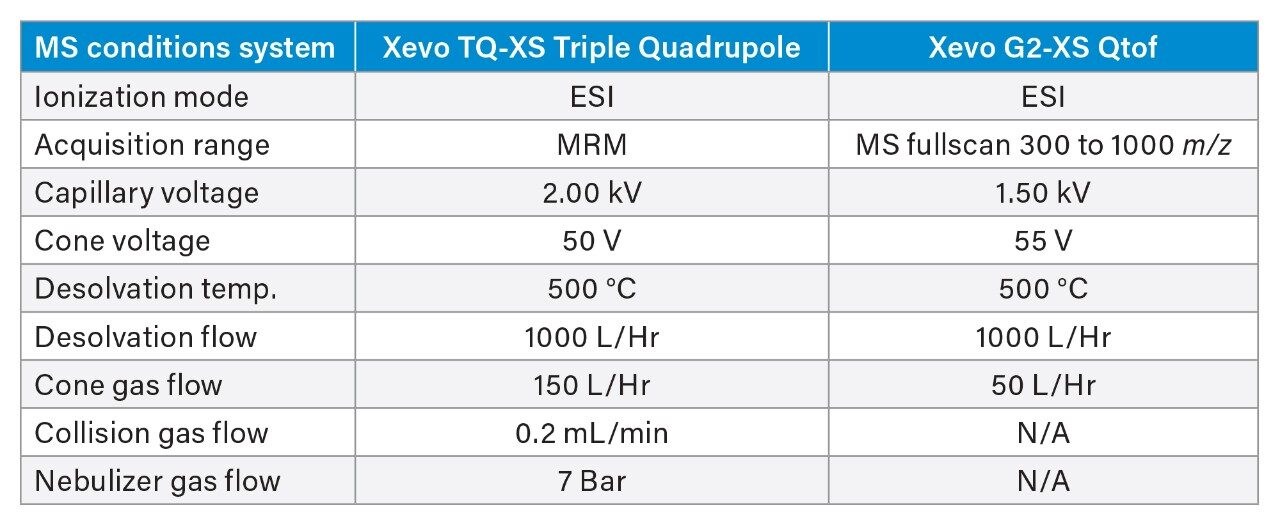
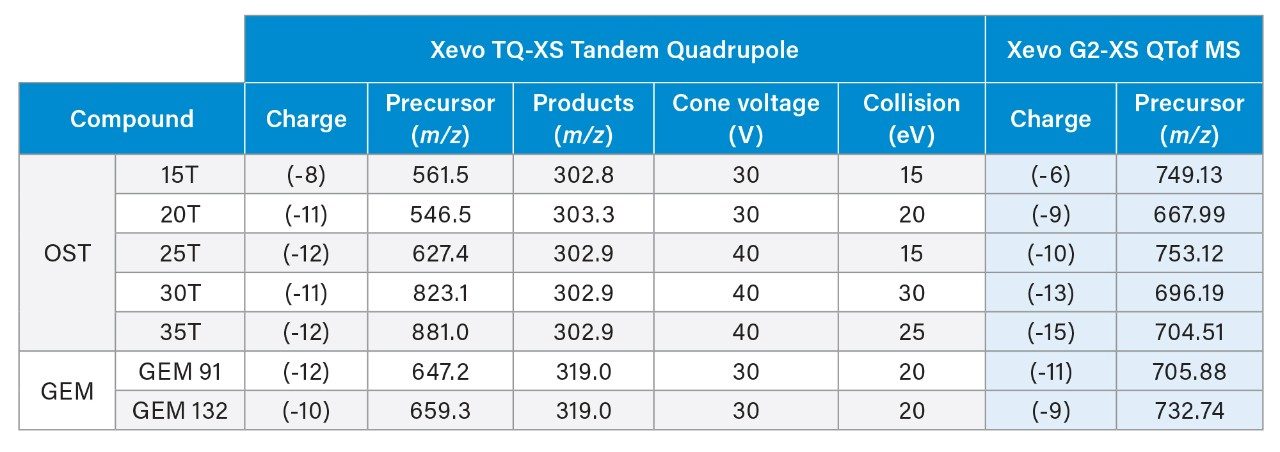
MS precursor and fragments used for the HRMS and MRM methods are shown in Table 1 (charges and m/z as shown in Table 1 for OST standard and GEM 91). For HRMS analysis, target enhancement was applied to m/z 700, increasing sensitivity for a 50-100 Da mass range around the mass of interest. For each platform, the most intense/selective charge state clusters were used and provided modest performance benefits to each platform. In general, two or three charge state clusters out of the charge envelope were found to have high abundance and could be selected depending on the nature and interference of background ions.
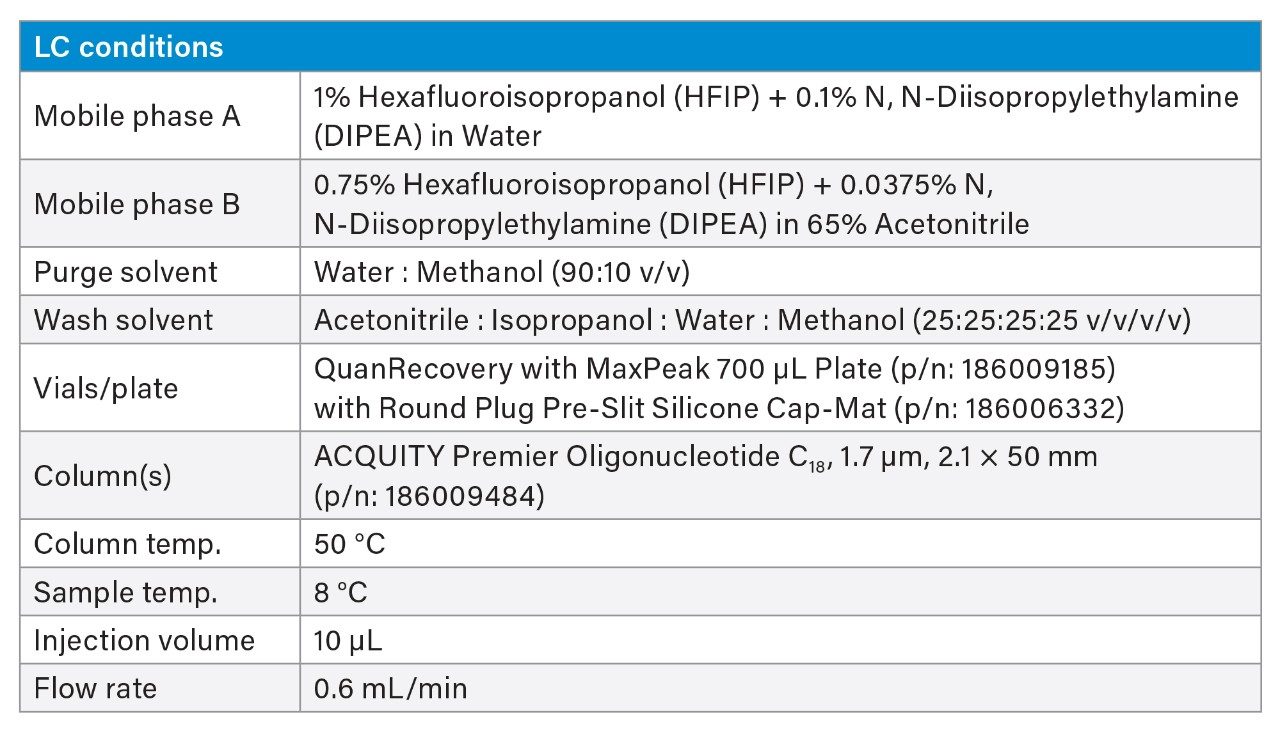
Chromatographic separation was achieved using an ACQUITY Premier BSM System, equipped with an ACQUITY Premier Oligonucleotide C18 Column, 130 Å, 1.7 µm, 2.1 x 50 mm (p/n: 186009484), using a 5-minute gradient (5–90% B) with 1% Hexafluoroisopropanol (HFIP) + 0.1% N, N-Diisopropylethylamine (DIPEA) in water as mobile phase A and 0.75% Hexafluoroisopropanol (HFIP) + 0.0375% N, N-Diisopropylethylamine (DIPEA) in 65% Acetonitrile as mobile phase B (flow rate 0.6 mL/min). ACQUITY Premier Columns incorporating MaxPeak High Performance Surfaces (HPS) Technology minimized non-specific bindingand improved recovery and assay limits of detection. HPS Technology was developed specifically to minimize metal interactions with analytes such as oligonucleotides, phosphopeptides, small molecule organophosphates, and other analytes that have historically shown strong affinity towards metal surfaces.2
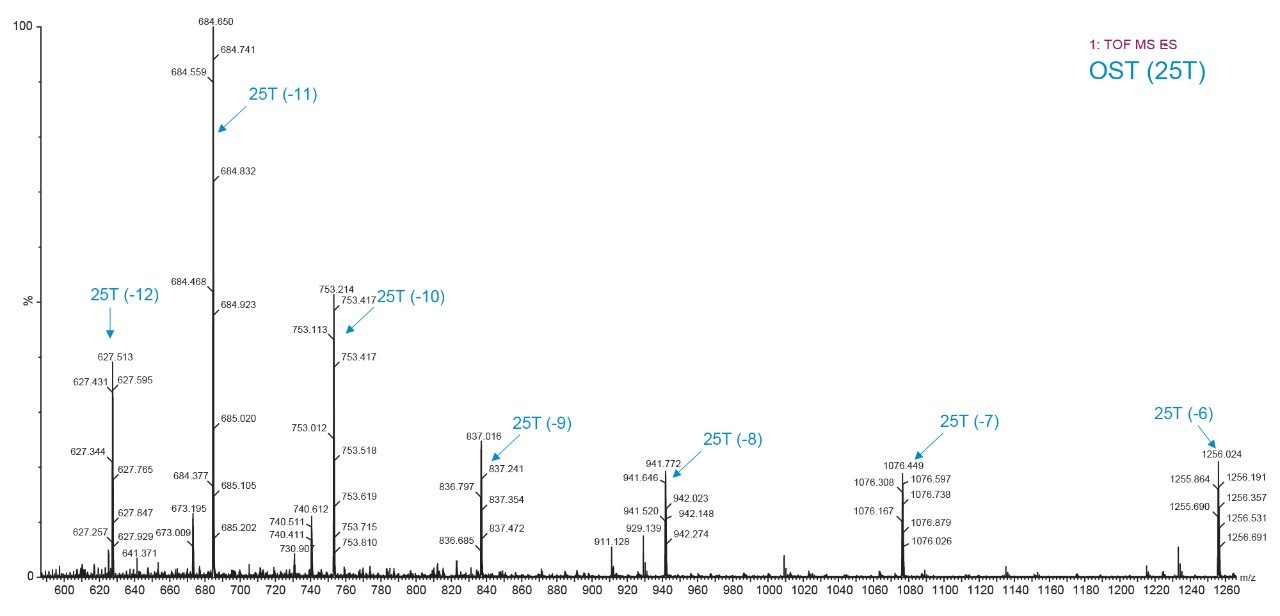
Figure 2 to Figure 4 show typical spectra observed for these compounds, showing both a general distribution of charge state clusters (Figure 2) and close-up spectra for charge states selected for monitoring (Figure 3, 4). HRMS enables characterization (identifying different isotopes/charge states) of an oligonucleotide molecule and the ability to select isotope/charge state best suited to achieve good sensitivity, selectivity, and accuracy for quantitation. During method development, multiple ions may be monitored. If a preferred ion is interfered with, a different cluster or isotope within a cluster may be easily chosen. Similarly, multiple ions may be quantitated simultaneously or monitored for interference. Finally, and perhaps most importantly, impurities, metabolites, and adducting may be readily observed and provide more information into both the quality of the assay, as well as additional information about potential interferences and changes in the samples themselves.
![Close-up view of the [M-10H]-10 charge state cluster used for quantitation of OST (25T) in this experiment](/content/dam/waters/en/app-notes/2021/720007391/720007391en-f3.jpg.82.resize/img.jpg)
![Close-up view of the [M-11H]-11 charge state cluster used for quantitation of GEM 91 in this experiment](/content/dam/waters/en/app-notes/2021/720007391/720007391en-f4.jpg.82.resize/img.jpg)
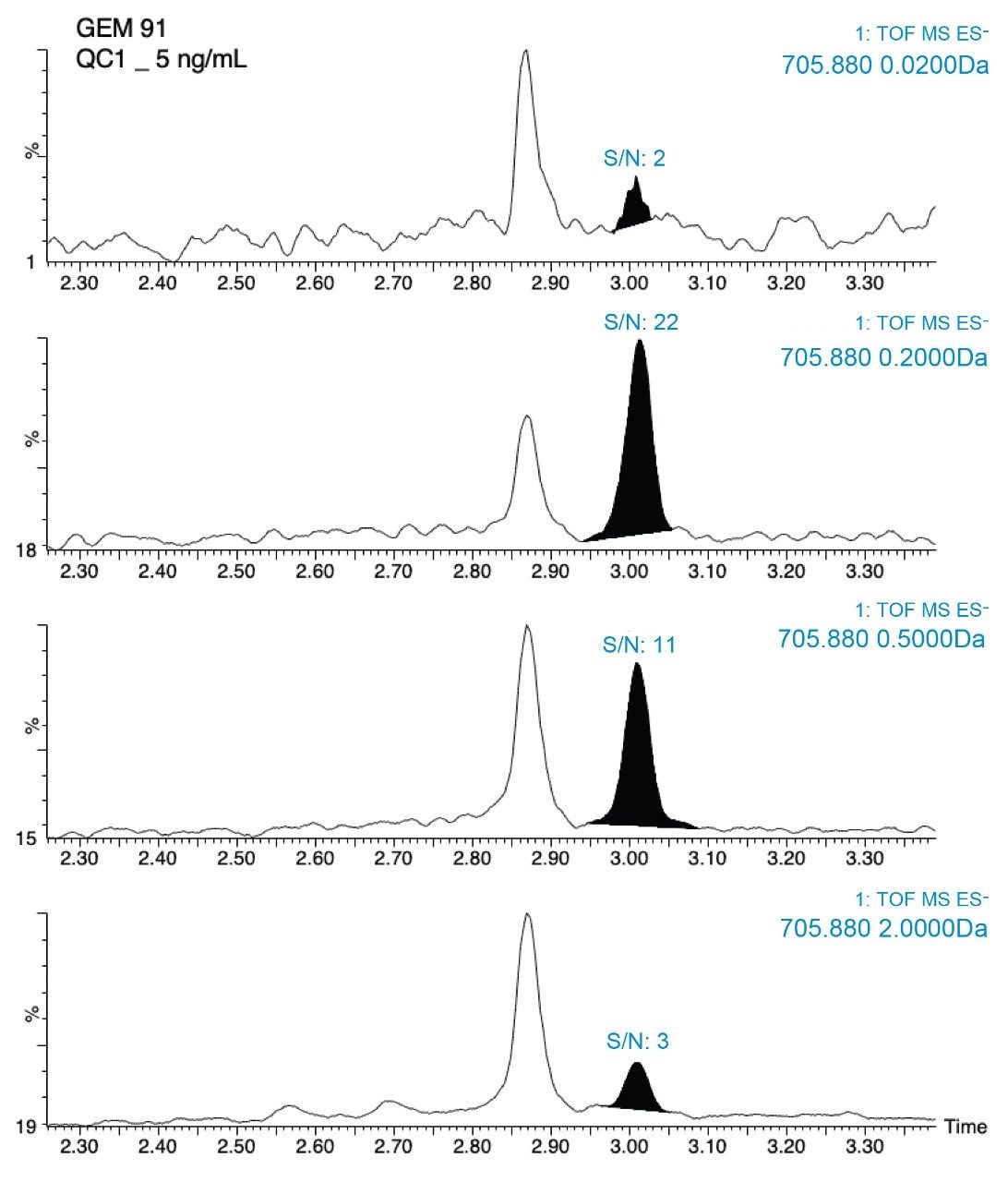
Figures 5a and 5b demonstrates the advantage of the high mass accuracy of HRMS for analysis of oligonucleotides to extract narrow mass windows, thereby greatly improving the selectivity of the assay. As seen in this figure, using a mass extraction window of 0.2 Da provided the best signal-to-noise, S/N and results in the best overall sensitivity. The QTof gives the user the ability to choose the mass extraction window (MEW) appropriate for the assay and fine tune selectivity. Figure 5 shows that in general, 0.2 Da was a good starting point for these analytes, providing the best S/N.
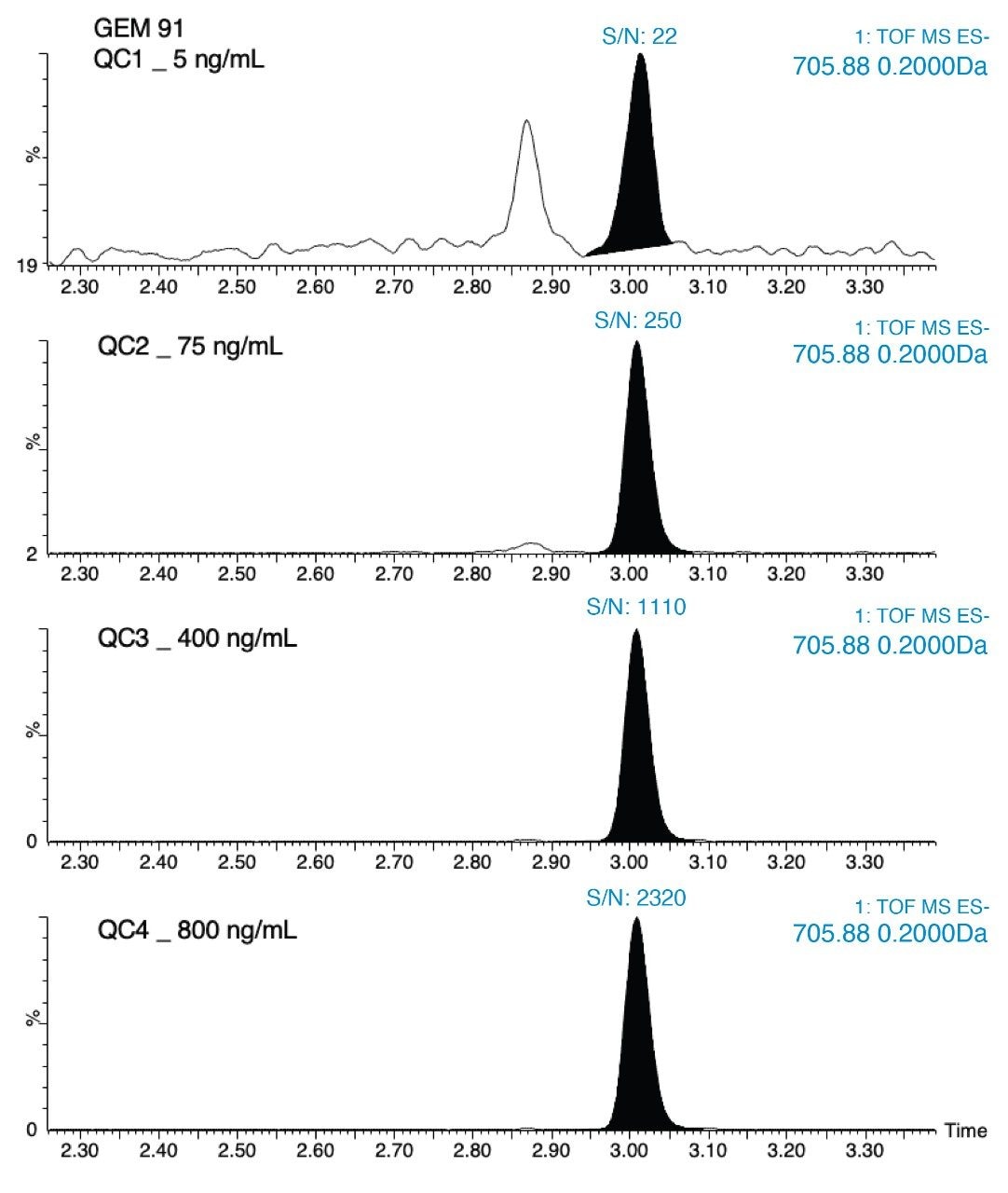
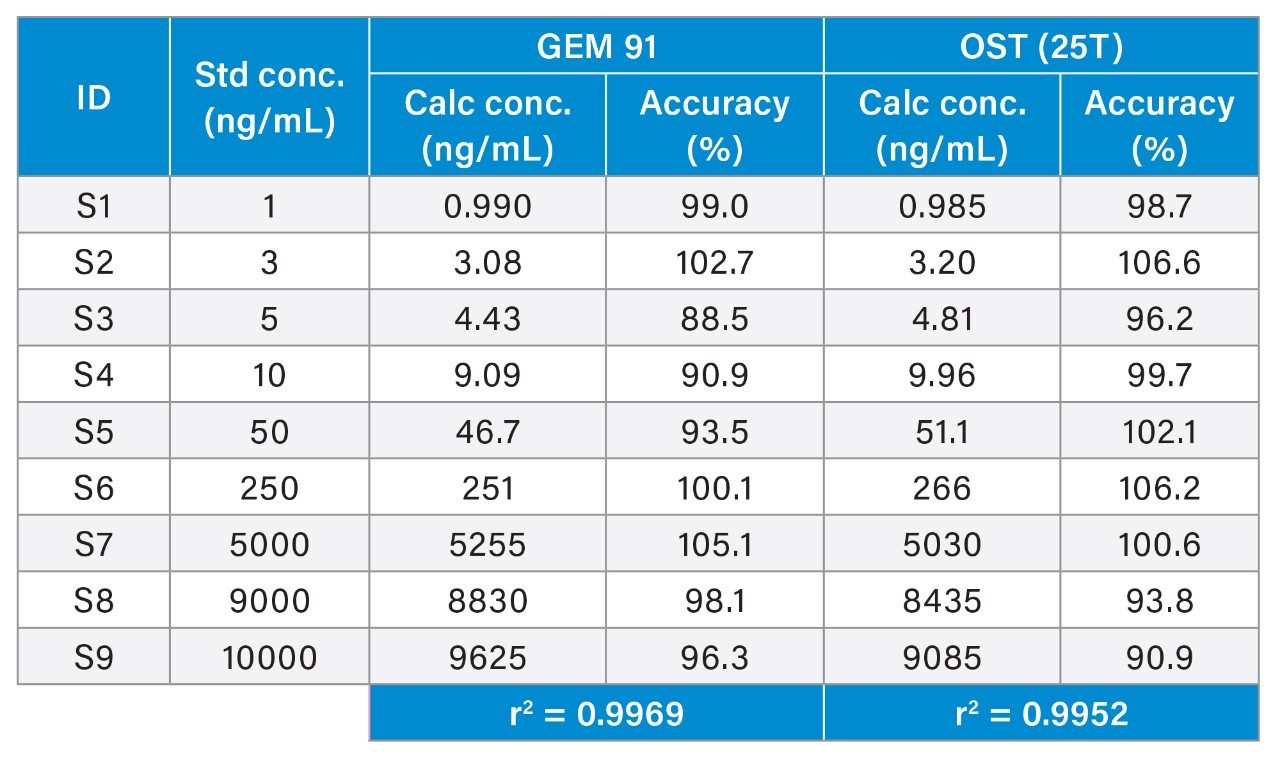
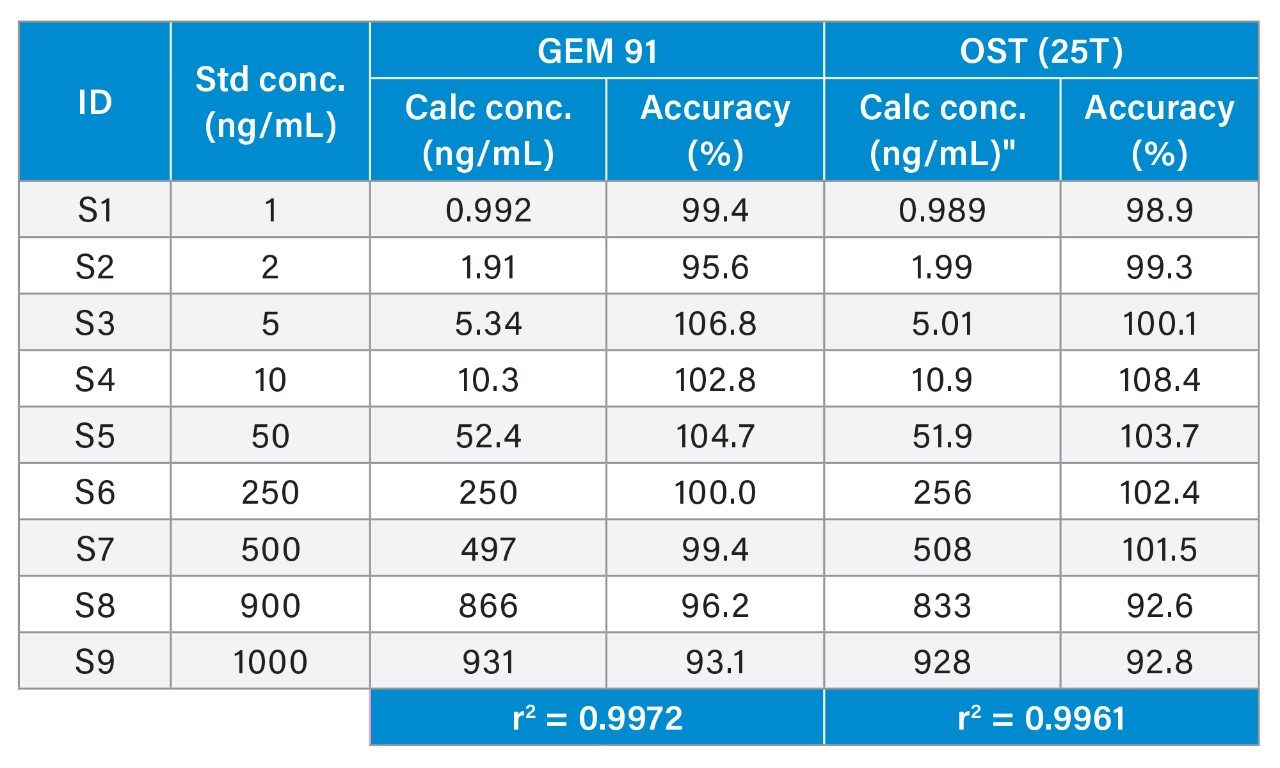
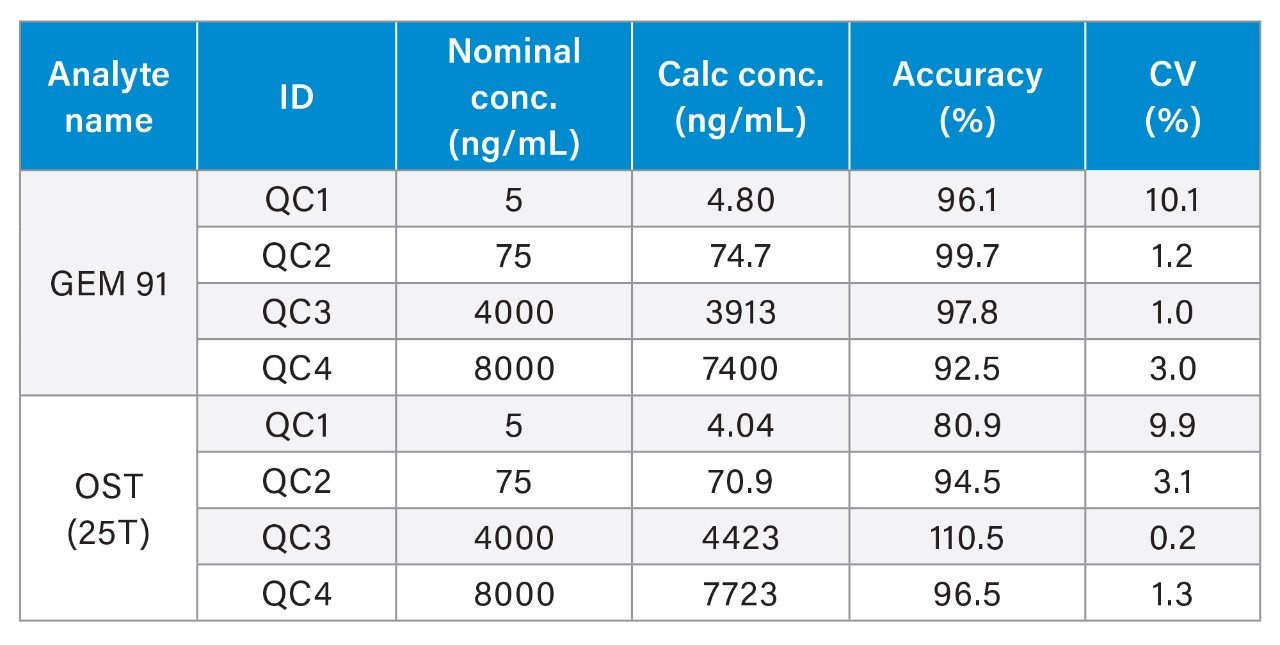
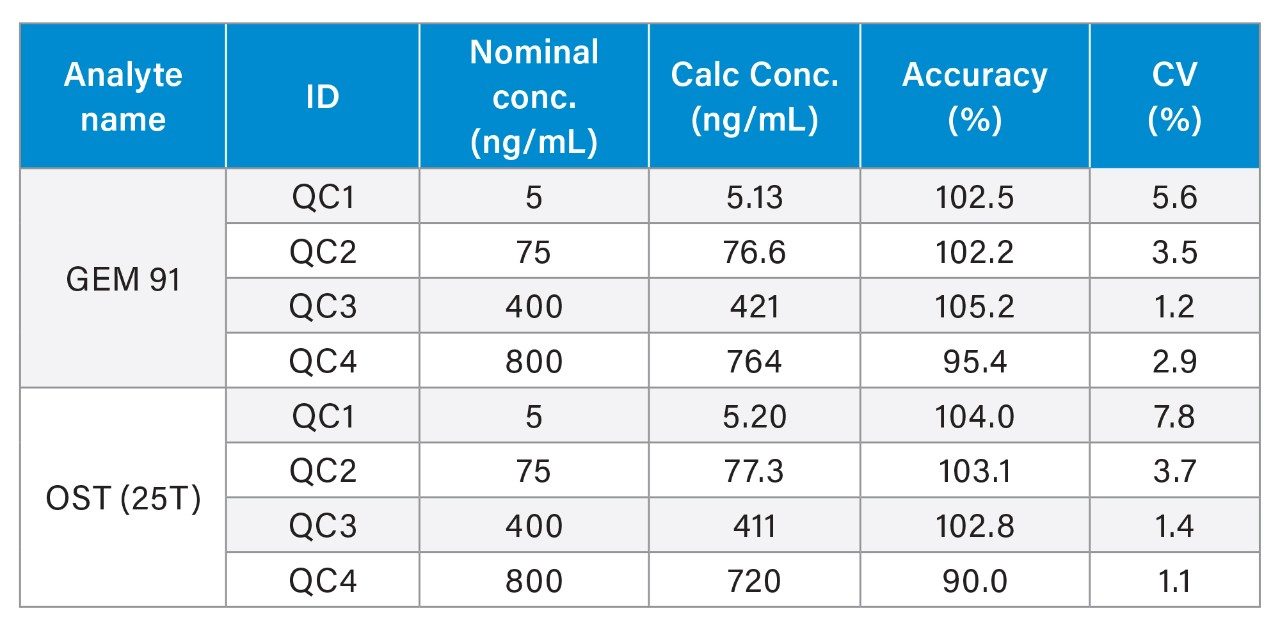
Overall, HRMS performance was comparable to MRM quantification data observed on the Xevo TQ-XS Triple Quadruole. A summary of standard curve performance for both OST (25T) and GEM 91 using HRMS or MRM analysis is shown in Tables 2 to 5. On both mass spectrometers, the lower limit of quantification (LLOQ) achieved was 1 ng/mL with a dynamic range from 1–1,000 ng/mL on the QTof and 1–10,000 ng/mL on the tandem MS. The calibration curves were linear with r2 values >0.99 (1/x2 weighting) with mean accuracy of all calibration points between 88–108%. Quality Control (QC) performance of Oligonucleotides OST and GEM 91 are reported in table X, with mean accuracies between 81–110% and CV’s between 0.2–10.1%.
Conclusion
- The Xevo G2-XS QTof MS has been proven to be a sensitive platform able to generate high quality data for LC-MS-based quantitation with multiple acquisition modes, providing multiple options for quantifiable peaks. It enables the ability to fine tune selectivity with the MEW and to select isotopes/charge states appropriate for the assay.
- Data (Tables 2 to 5) shows both Xevo G2-XS QTof MS and Xevo TQ-XS Triple Quadrupole MS can generate 1 ng/mL levels of sensitivity for oligonucleotides without compromising linearity, accuracy and precision.
- ACQUITY Premier LC System equipped with an ACQUITY Premier Oligonucleotide C18 Column are essential tools for improving recovery and assay limits of detection for challenging analytes.
- The use of ACQUITY Premier technologies helps to mitigate metal adsorption, ensuring robust and sensitive quantitation performance for quantitative bioanalytical assays.
References
- Jaeah Kim, Babak Basiri, Chopie Hassan, Carine Punt, Erik van der Hage, Cathaline den Besten, Michael G. Bartlett, Metabolite Profiling of the Antisense Oligonucleotide Eluforsen Using Liquid Chromatography-Mass Spectrometry, Molecular Therapy – Nucleic Acids, Volume 17, P714–725, September 6, 2019.
- Kathryn Brennan, Mary Trudeau, Paul D. Rainville, Utilization of the ACQUITY Premier System and Column for Improved Oligonucleotide Bioanalytical Chromatographic Performance, Waters Application Note, 720007119, January 2021.
- Kathryn Brennan, Mary Trudeau, Michael Donegan, Paul D. Rainville, Improved Oligonucleotide SPE-LC-MS Analysis Using MaxPeak High Performance Technology, Waters Application Note, 720007019, September 2020.
- Jennifer M Nguyen, Martin Gilar, Brooke Koshel, Michael Donegan, Jason MacLean, Zhimin Li, Matthew A Lauber, Assessing the Impact of Nonspecific Binding on Oligonucleotide Bioanalysis, Bioanalysis, Vol. 13, No. 16, Pages:1233–1244.
720007391, October 2021