Use of Predicted Versus Measured CCS Values from Different Instrument Platforms, and Isomer Separation on the SELECT SERIES Cyclic IMS
Abstract
Biotransformation activities require the comparison of metabolites across species and studies. In general, chromatographic retention time, accurate mass measurement, and mass spectral data are used to align metabolites. Isomeric metabolite comparison may be more challenging particularly when retention times may differ depending on the analytical conditions used. Additionally, the elemental formulae as well as tandem mass spectrometry (MS/MS) spectra can be identical which significantly increases the complexity of the data interpretation and localization of the biotransformation. The use of collision cross section (CCS) values to compare metabolites analyzed using the SELECT SERIES Cyclic IMS and the SYNAPT G2-Si QTof instruments located in different facilities has been shown here and demonstrates the benefit of this analyte-specific physiochemical property to align metabolites across studies.
Moreover, computational prediction of CCS values may provide an additional data asset, allowing the comparison of predicted with measured CCS values. This can further provide additional insights to differentiate between isomers. The prediction can also be used to suggest when additional cyclic ion mobility separation (cIMS) would be beneficial in the separation of isomers and increase confidence in any assignment with the use of higher ion mobility resolution. Examples are given here where cIMS has been used to separate oxygenated metabolites of ranitidine and imipramine; this alternative separation mechanism adds to the separating power of UltraPerformance Liquid Chromatography (UPLC) and is of benefit when isomers co-elute.
Benefits
- Leverage flexible resourcing as CCS values enable metabolite tracking across studies, species, and facilities using ion mobility-enabled instrumentation
- CCSOnDemand predicts CCS values, and it shows potential for assisting the structural elucidation of metabolites
- Differentiate isomeric metabolites with enhanced ion mobility resolution provided by cIMS
Introduction
In vitro and in vivo metabolism studies are key elements during drug development. Typically, the level of complexity and study detail increases alongside drug development to address safety aspects with regards to human-specific or disproportionate metabolites and to identify metabolites contributing to pharmacological activity.
At the discovery stage, generic high-throughput liquid chromatography-mass spectrometry (LC-MS) methods with short gradients are used, focusing mainly on major metabolites to identify molecular liabilities within the potential drug candidate. In contrast, dedicated LC-MS methods with long gradients are used at development stage to provide chromatographic separation and structural elucidation of all observed metabolites to ensure safety coverage of any human metabolite in animal species as requested by health authorities.1,2
Conventionally, retention times and mass spectral data have been used to identify metabolites across species and studies. However, this can be problematic for closely eluting isomeric metabolites particularly if their MS/MS spectra are indistinguishable. Moreover, the order of elution may change depending on the chromatographic conditions used either internally or at an external service provider. Ion mobility-mass spectrometry (IMS) adds another analytical dimension, enabling determination of the molecules’ CCS value which is calculated from its drift time. This physiochemical property is analyte-specific and represents a robust parameter, allowing the tracking of metabolites across multiple platforms, various conditions, matrices, and studies as it is unaffected by these changes. In addition, the prediction of CCS values via computational techniques such as machine-learning or quantum mechanics is of great interest to aid in the structural identification of metabolites and derive their metabolic pathways.
Here, we compare CCS values for a series of approved drugs and their metabolites obtained using two different IMS enabled mass spectrometers (the SELECT SERIES Cyclic IMS and the SYNAPT G2-Si QTof), which were located in different facilities and operated by different scientists. Moreover, CCSonDemand,3 a machine-learning algorithm, was used to predict CCS values and compare theoretical with measured CCS values from each instrument. The cIMS technology was further used, providing increased IMS resolution, to distinguish isomeric metabolites, which was not possible on a commercially available linear IMS device.
Experimental
Sample Description
Reference standards of approved drugs and their metabolites (a total of 26 compounds) were first dissolved in dimethyl sulfoxide to obtain stock solutions at 1 mM. Working solutions at 10 µM were subsequently prepared for each compound following dilution of each stock solution in acetonitrile/water (1/1, v/v), which were then used for analysis.
Method Conditions
Predicted CCS values were obtained using CCSOnDemand, an experimental predictive machine-learning based algorithm.
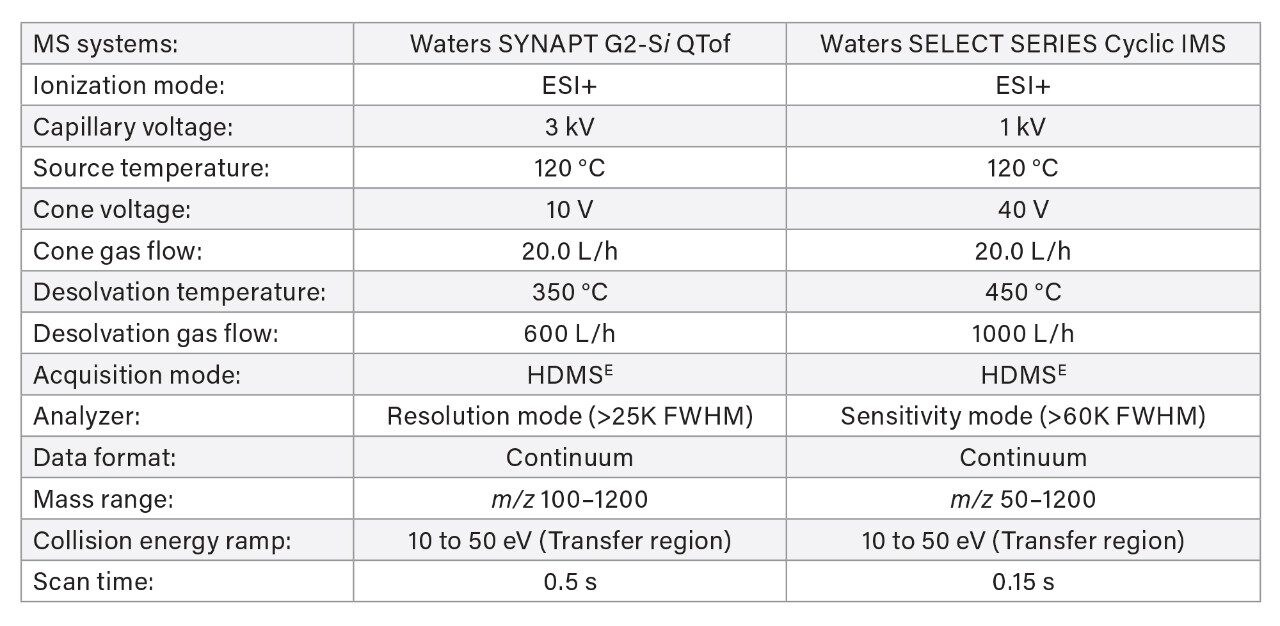
Measured CCS values were determined following HDMSE analysis (n=3) either performed on a SYNAPT G2-Si QTof at Novartis (Basel, Switzerland) or a SELECT SERIES Cyclic IMS (single pass) at Waters (Wilmslow, UK). Corresponding LC-MS conditions are summarized below. In both cases, data acquisition was conducted with MassLynx (v4.2) whereas data processing was performed with UNIFI (v1.9.4) to determine measured CCS values. Arrival time distribution (ATD) plots were obtained using DriftScope (v3.0) and MassLynx (v4.2).
To further demonstrate the ability of cIMS to separate isomeric metabolites, mixtures of isomeric metabolites were infused into the ion source at 5 µL/min and data were acquired using single and multiple passes.
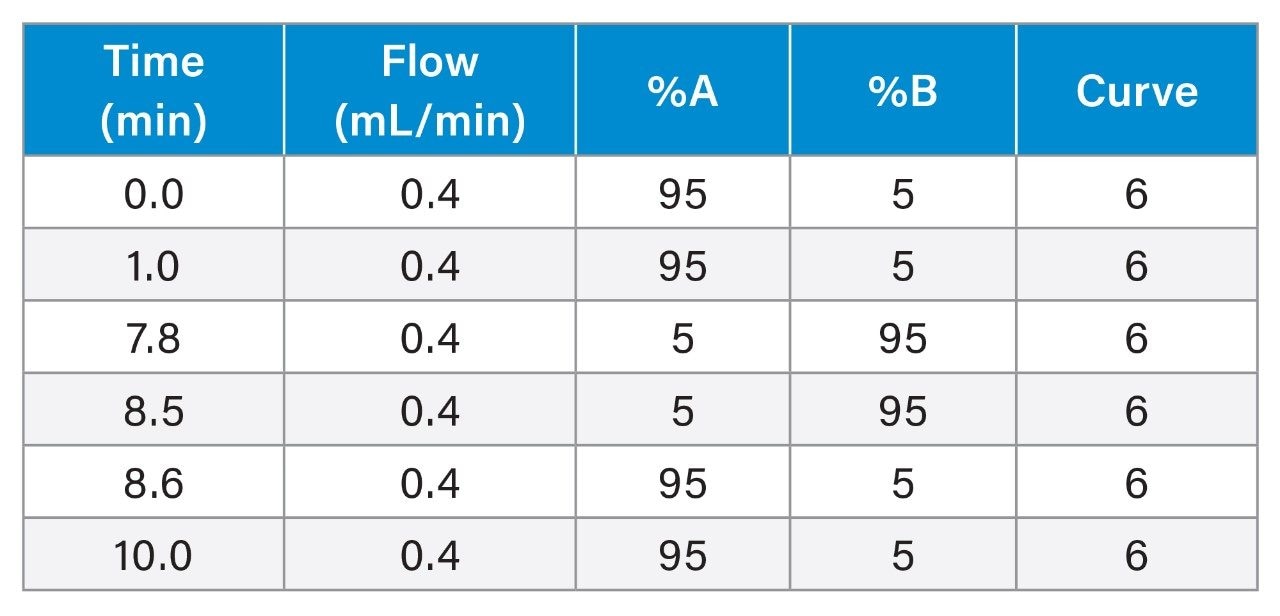
UPLC Conditions
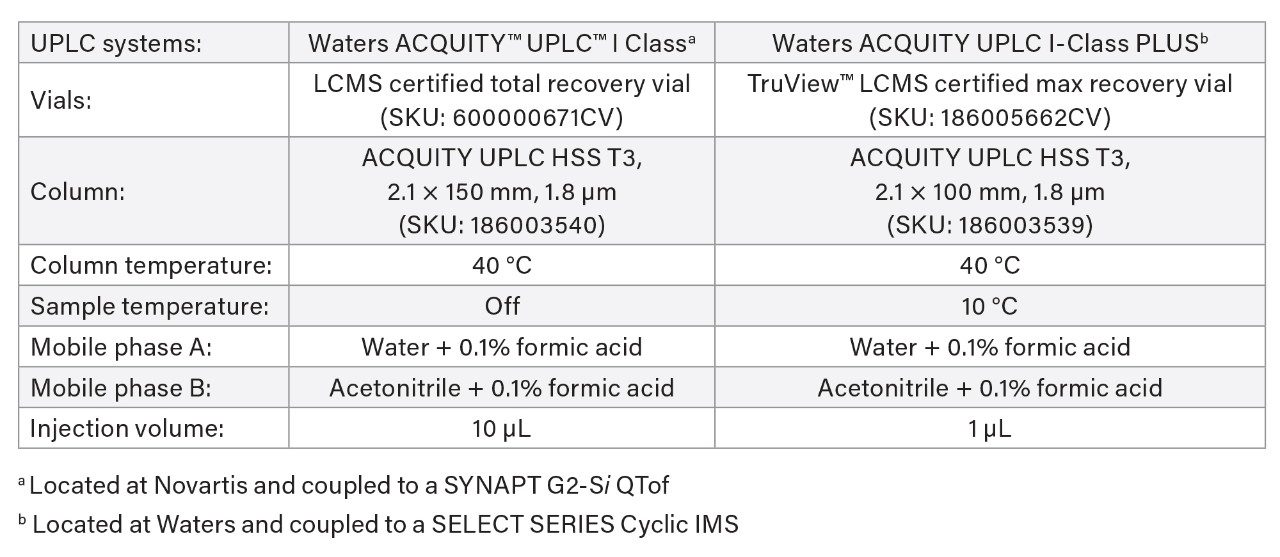
Gradient Table
MS Conditions
Results and Discussion
Following triplicate analysis of a total of 26 compounds either on the SYNAPT G2-Si QTof or SELECT SERIES Cyclic IMS with MassLynx data acquisition, the HDMSE data were imported into UNIFI to determine their CCS values (Table 1). The root mean square error (RMSE) across all these measurements was 1.0% showing excellent agreement between the two data sets.
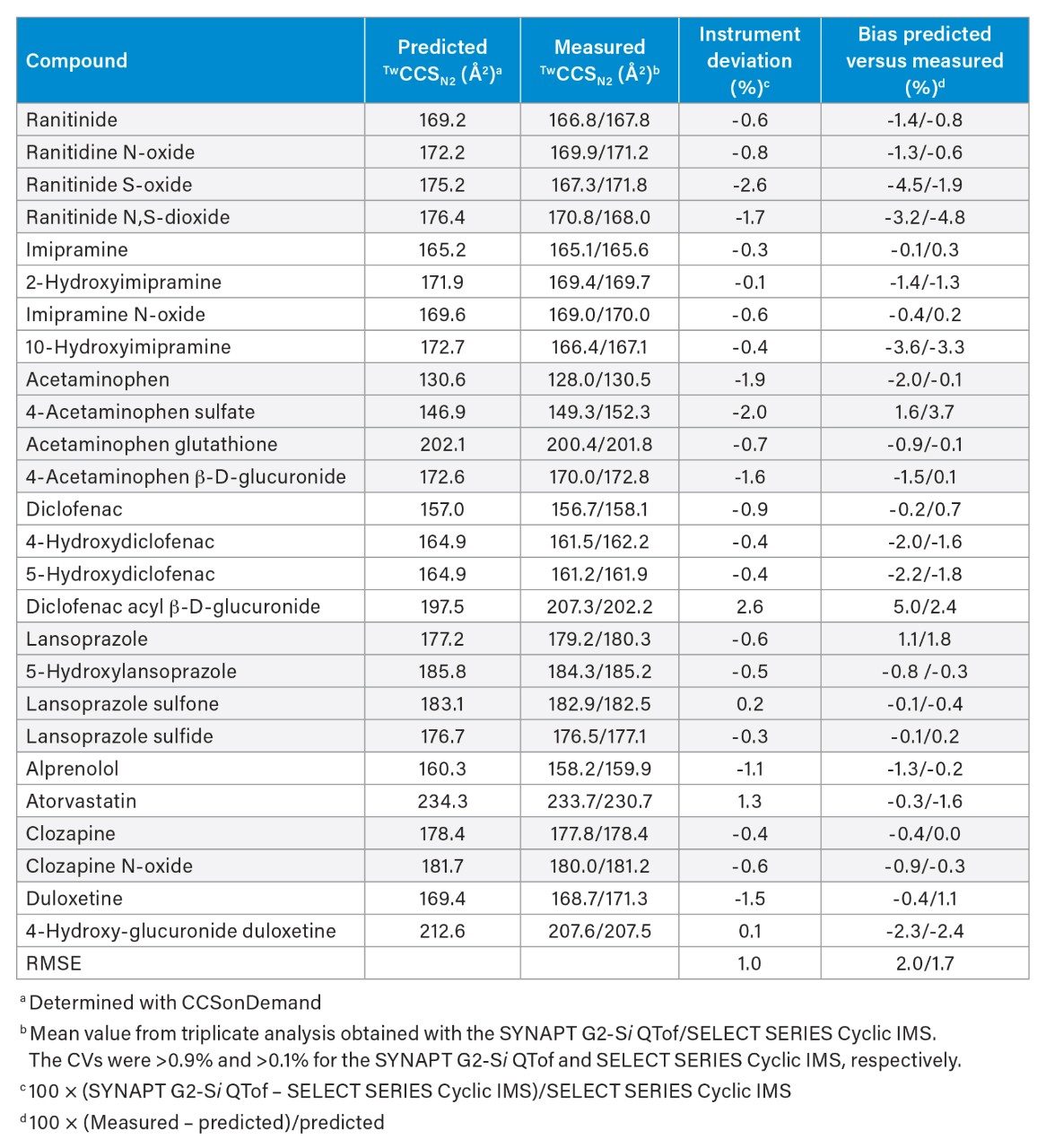
This excellent agreement in CCS values across both platforms for a wide range of approved drugs including their metabolites demonstrated the benefit of CCS values in addition to retention time and mass spectral data to track metabolites across different studies, methods, and facilities. The CCS value can be particularly useful when product ion mass spectra observed for isomeric metabolites cannot be differentiated4,5 and when retention times differ due to extended separation of new metabolites6 or if LC conditions/equipment have been changed during drug development.
In addition, CCS values were predicted for all compounds using CCSOnDemand. The difference between predicted and mean measured CCS values was within ±5.0% for all 26 compounds regardless of the platform used. The calculated RMSE for the bias of 2.0% and 1.7% confirmed the ability to predict CCS values for certain metabolites. These predictions would not only aid in structure elucidation and better assign the localization of the biotransformation, but also allow the characterization of metabolic pathways more efficiently.
Moreover, the predicted CCS value can also give an indication of whether isomeric metabolites can be separated by ion mobility. For example, the predicted CCS value for ranitidine N- and S-oxide differed by 3 Å2 as confirmed by the mean measured CCS values (Table 1). Assuming both isomeric metabolites would co-elute chromatographically and identical mass spectral as well as linear IMS data were obtained (similar to a single pass experiment with cIMS), the conclusion would have been that only one isomer existed. However, by extending the number of passes during cIMS analysis, which significantly increases IMS resolution,7 both isomers could be separated as indicated in the corresponding ATD plots (Figure 1).
![ATD plots of m/z 331.1440 [M+H]+ for ranitidine N- and S-oxide following 1 to 4 passes during cIMS analysis](/content/dam/waters/en/app-notes/2022/720007514/720007514en-f1.jpg.82.resize/img.jpg)
A similar situation was obtained with the oxygenated metabolites of imipramine (Figure 2). The predicted CCS for 2-hydroxyimipramine, imipramine N-oxide, and 10-hydroxyimipramine suggested that these isomeric metabolites may be separated using ion mobility. After single pass, the three isomers were not separated. However, 10-hydroxyimipramine could be observed as a shoulder following two passes with almost baseline resolution after four passes. The remaining two oxygenated metabolites could not be distinguished until two maxima were seen following 13 passes. To provide separation of the 2-hydroxyimipramine and imipramine N-oxide, 10-hydroxyimipramine was ejected from the cyclic array and the other two isomeric metabolites were separated by cIMS following 25 passes, resulting in a resolution of 325 (Ω/∆Ω).
![ATD plots of m/z 297.1967 [M+H]+ for oxygenated imipramine metabolites following 1 to 4, 13, and 25* passes during cIMS analysis](/content/dam/waters/en/app-notes/2022/720007514/720007514en-f2.jpg.82.resize/img.jpg)
In general, the separation times used following multiple passes are compatible with UPLC separation as drift times after four passes are less than 100 ms and the loss in transmission with each pass (about 2.4%8) has minimal impact on the data. These multiple pass experiments can therefore be used in combination with UPLC to provide enhanced resolution over that provided by mass resolution alone.
Conclusion
Regardless of the IMS platform (SYNAPT G2-Si QTof or SELECT SERIES Cyclic IMS), mean measured CCS values were identical and in agreement with predicted CCS values using the CCSonDemand machine-learning algorithm. Since CCS values are robust and analyte-specific, this physiochemical parameter can assist biotransformation scientists during drug discovery and development stage where various studies, facilities, and utilized analytical methods are involved. In particular, the usage of CCS values during metabolism studies will ease structural elucidation of identified isomeric metabolites-exhibiting similar retention times and mass spectral data. Moreover, metabolic pathways can be derived more efficiently by assigning the origin of metabolites based on relative CCS values, providing greater confidence in generated metabolism data.
Finally, accurately predicted CCS values act as an indicator to whether higher ion mobility resolution is required to separate isomeric metabolites. Here we demonstrated the need for an ion mobility resolution of up to 325 (Ω/∆Ω) by using the unique multipass cyclic ion mobility capability of the SELECT SERIES Cyclic IMS for the separation of several isomeric metabolites throughout this study.
Acknowledgements
The authors highly acknowledge the support from Markus Walles (Novartis), Mario Mergelsberg (Waters), and Emma Harry (Waters).
David Higton - Waters Corporation
Christian Lanshoeft, Frederic Lozac'h - Novartis Pharma AG.
References
- ICH M3 (R2) Non-clinical Safety Studies for the Conduct of Human Clinical Trials for Pharmaceuticals, 2013.
- FDA Guidance for Industry. Safety Testing of Drug Metabolites, 2020.
- Broeckling C, Yao L, Isaac G, Gioioso M, Ianchis V, Vissers JPC. Application of Predicted Collisional Cross Section to Metabolome Databases to Probabilistically Describe the Current and Future Ion Mobility Mass Spectrometry. J Am Soc Mass Spectrom, 2021, 32, 661–669.
- Higton D, Palmer ME, Vissers JPC, Mullin LG, Plumb RS, Wilson ID. Use of Cyclic Ion Mobility Spectrometry (cIM)-Mass Spectrometry to Study the Intramolecular Transacylation of Diclofenac Acyl Glucuronide. Anal Chem, 2021, 93, 20, 7413–7421.
- Connolly JFRB, Munoz-Muriedas J, Lapthorn C, Higton D, Vissers JPC, Webb A, Beaumont C, Dear G.J. Investigation into Small Molecule Isomeric Glucuronide Metabolite Differentiation Using In Silico and Experimental Collision Cross-Section Values. J. Am. Soc. Mass Spectrom. 2021; 32 (8): 1976–1986.
- Holdsworth C, Clayton R, Robinson H, Lord-Mears C, Kendrick J. Utilization of Ion Mobility Enabled Collisional Cross Section Measurements for the Comparison of Metabolites across Differing Chromatographic Methods. Poster DMDG 2016.
- Giles K, Ujma J, Wildgoose J, Pringle S, Richardson K, Langridge D, Green M. A Cyclic Ion Mobility-Mass Spectrometry System. Anal. Chem. 2019, 91, 13, 8564–8573.
720007514, February 2022